Screening assays for antimicrobial agents
a technology of antimicrobial agents and assays, applied in the field of drug discovery, can solve the problems of rapid growth of such pathogens, and achieve the effects of reducing the biological activity or expression level of drug resistance-confident, effective inhibiting the growth of drug-resistant microbial pathogens, and improving the ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Identification of Drug-Resistant RpoB Mutants
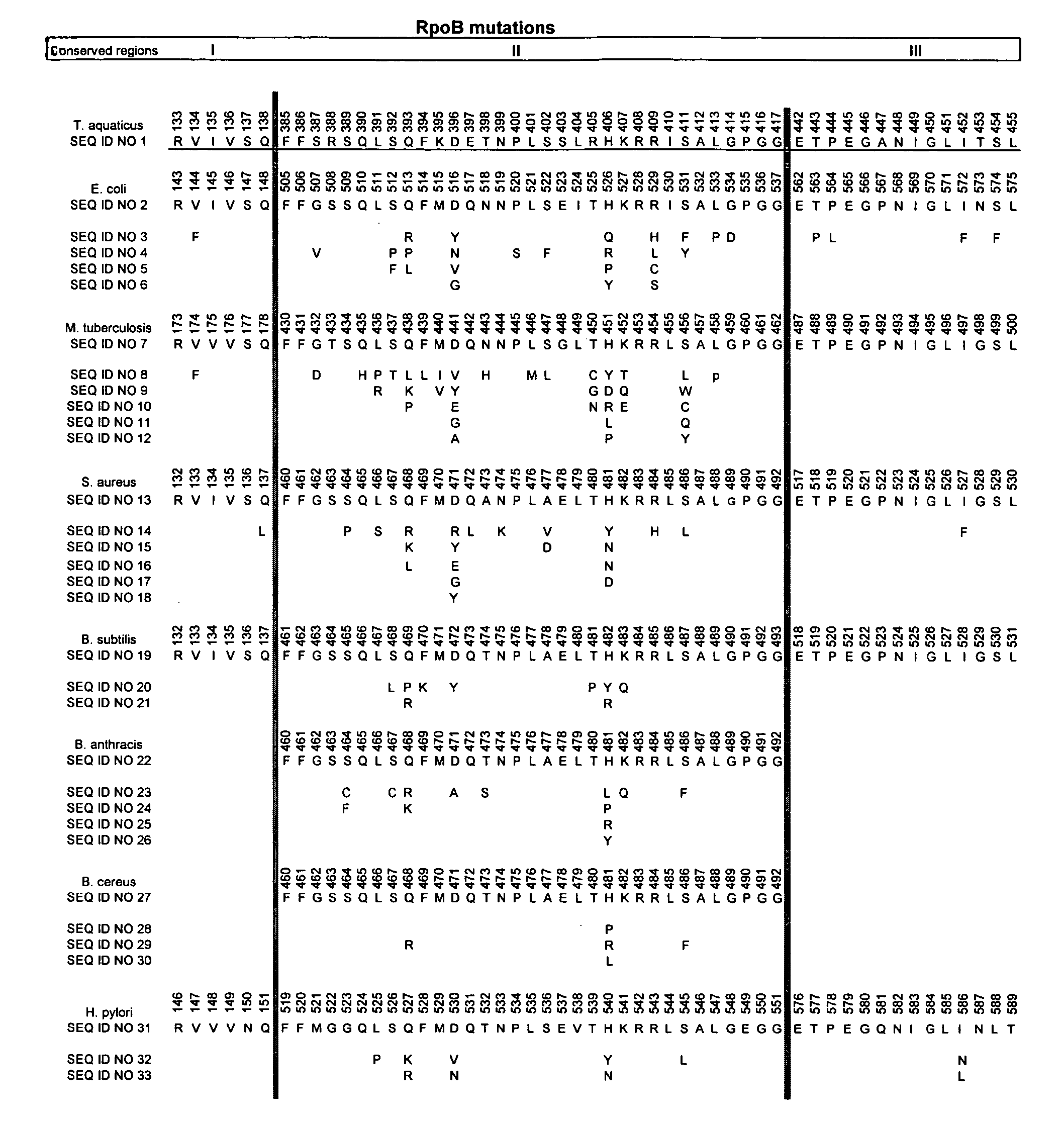
[0081]S. aureus is one of the most frequently encountered Gram-positive pathogens. Drug resistance-conferring mutations typically occur within the β subunit of RNA polymerase (RpoB mutations), in the rifampin-binding site. We have identified a collection of rifampin-resistant S. aureus mutants, as shown in Table 2. Mutants of S. aureus were isolated by inoculating a culture of S. aureus ATCC strain 29213 (standard susceptibility testing strain) into medium containing rifampin or rifalazil; by inoculating S. aureus ATCC strain 29213 into medium containing chemical derivatives of rifamycin (NCEs); by inoculating S. aureus Smith, a variant optimally adapted to colonizing and causing disease in the mouse septicemia model, into medium containing rifampin; and by using the Ian Chopra collection, the parent strain of which is S. aureus 8325-4 and described previously by Oliva et al. (Antimicrob. Agents Chemother. 45:532-9, 2001). Mutants resistant
example 2
In vitro Screening Using Mutant RpoB Polypeptides
[0086] Antimicrobial compounds may be identified by screening for interactions with mutated RpoB polypeptides in vitro. In one particular assay, various mutated RpoB polypeptides from S. aureus are immobilized in the wells of a multi-well plate such that each different mutant is present in its own well. A plurality of labeled candidate compounds are then individually contacted with each mutated RpoB polypeptide such that each candidate compound-polypeptide contacting event is segregated from the others. After a sufficient time to allow for binding, unbound compound is removed by washing and the presence of bound compound is determined by detection of the label. Candidate compounds that bind mutant RpoB polypeptides are thus identified.
[0087] In this example, compounds identified as binding RpoB are next optimally tested for their ability to reduce RNA polymerase activity. Each of the compounds identified as binding one or more RpoB pol
example 3
In vivo Screening of Rifamycin Derivatives
[0088] Antimicrobial compounds may also be screened in vivo using a mouse septicemia model. In one particular example, mice were inoculated with an S. aureus Smith strain (Weiss) encoding a mutated microbial polypeptide (e.g., a mutated RpoB). Compounds were administered either IV or orally 30 minutes following inoculation, and observations was continued for three days. Compounds that promoted the survival of inoculated mice were identified as being compounds that are effective against antibiotic-resistant forms of S. aureus.
[0089] In one example, when mutant strains derived from S. aureus Smith containing the RpoB H481Y alteration were inoculated in the mouse model by IV or oral route, two compounds, compound 15 and compound 16 from Table 3, were found to protect mice from lethality, relative to untreated animals (Table 4). The dose that was essential for efficacy was found to be considerably higher for the mutant strains than for the wild-t
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap