Compositions and methods for diagnosing and treating cancer
a cancer and composition technology, applied in the field of compositions and methods for diagnosing cancer, can solve the problems of increasing the quality of life, prolonging the disease-free state and overall survival rate, and reducing the risk of metastasis and death
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Identifying Stem Cell Cancer Markers
[0200]This Example describes how various stem cell cancer markers were identified using microarray screens. The results of these screens were processed and the names of the differentially expressed genes are reported in Tables 9A, 9B, 9C, 9D, 9E, 9F, 9G, 9H, 9I, 9J, 9K1, 9K2, 9L1, 9L2, 9M1, 9M2, 9N1 and 9N2; filed as Tables A, B, C, D, E, F, G, H, I, J, K1, K2, L1, L2, M1, M2, N1 and N2 in U.S. application Ser. No. 10 / 864,207, which is herein incorporated by reference.
[0201]In order to generate gene expression profiles, human breast tumorigenic cells that were initially isolated. A series of samples were accumulated from human breast tumors or normal tissues. These were generated as follows. Three passaged breast tumors—breast tumor cells from patient 1, 2, 3 were engrafted on mice. Each tumor was engrafted on three mice to make the triplicate tumors. The breast tumorigenic cells were then isolated from these tumors. Two or three unpassaged breast
example 2
Identification of Cancer Stem Cell Gene Signatures that Predict Clinical Outcome
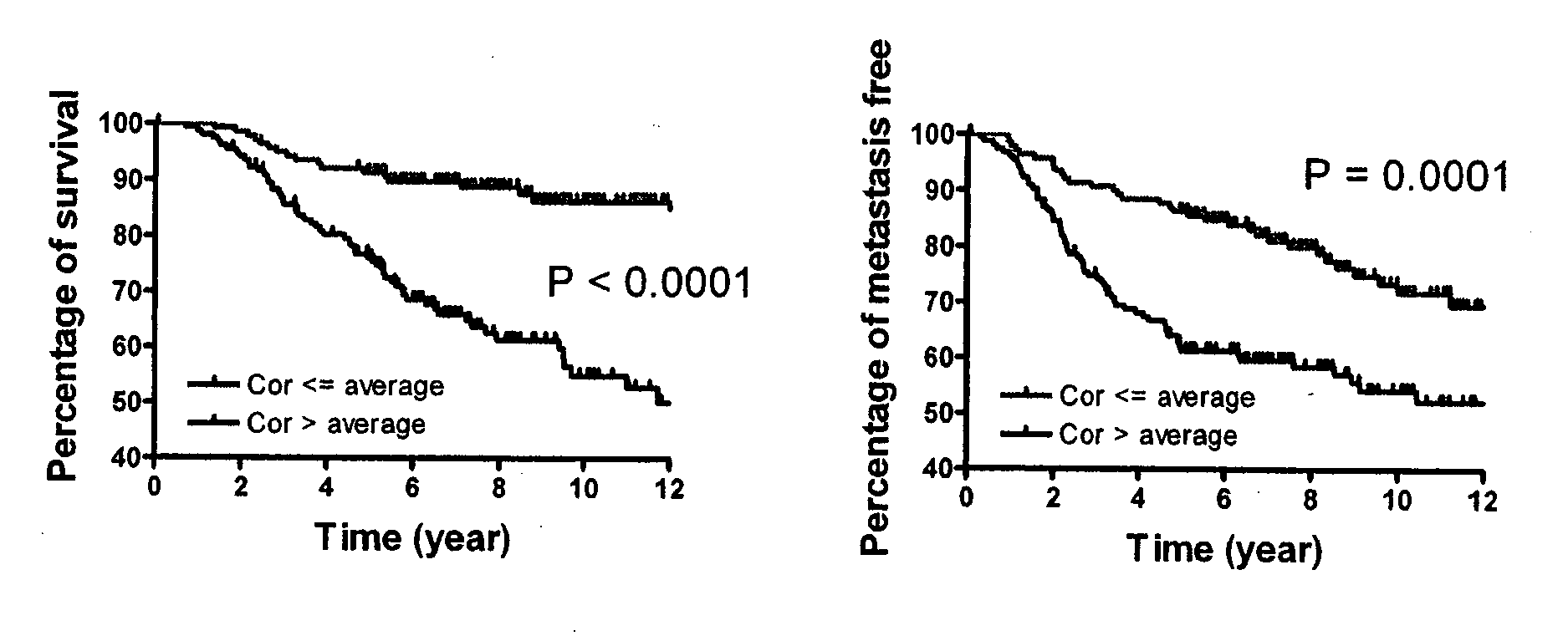
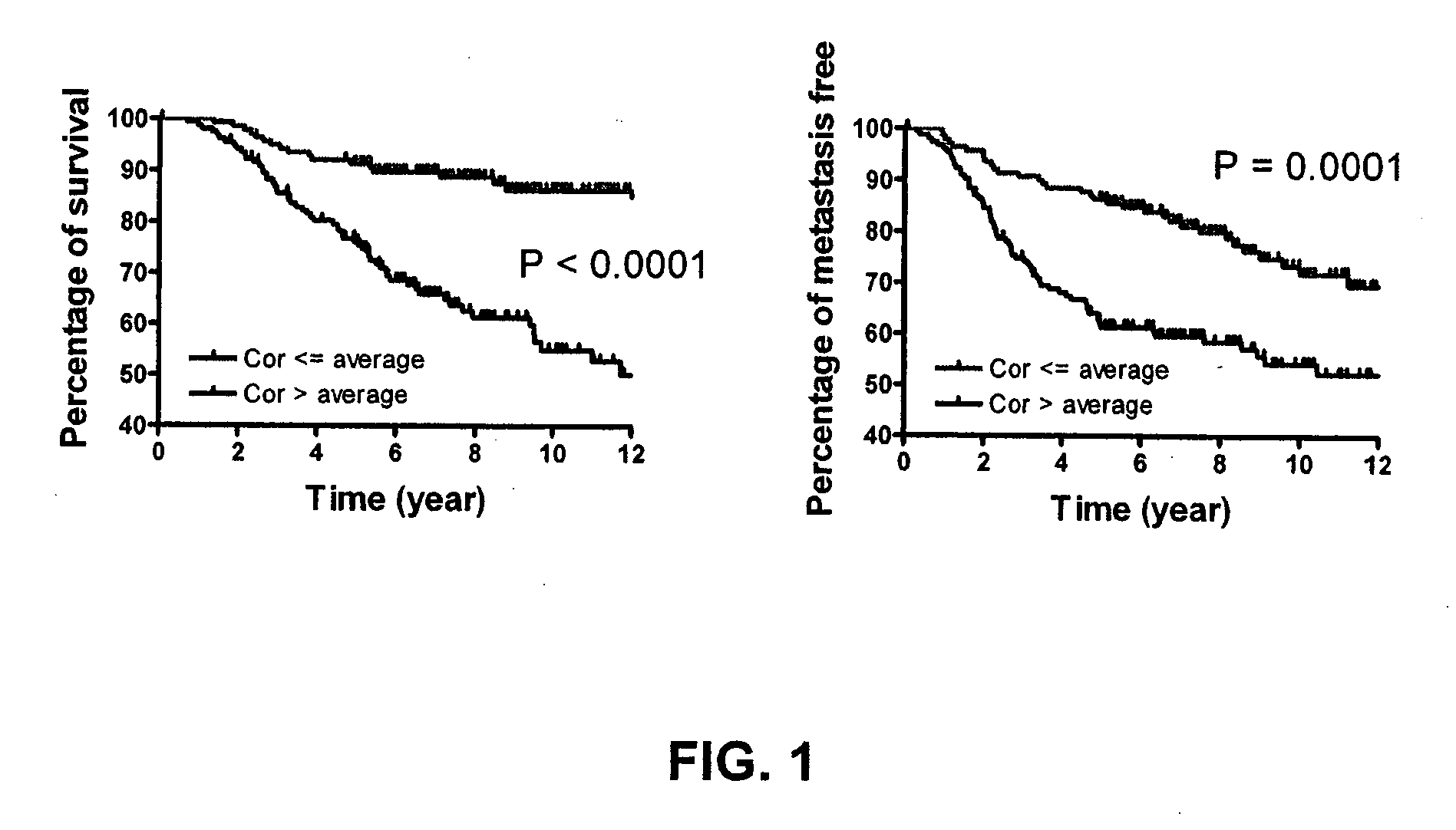
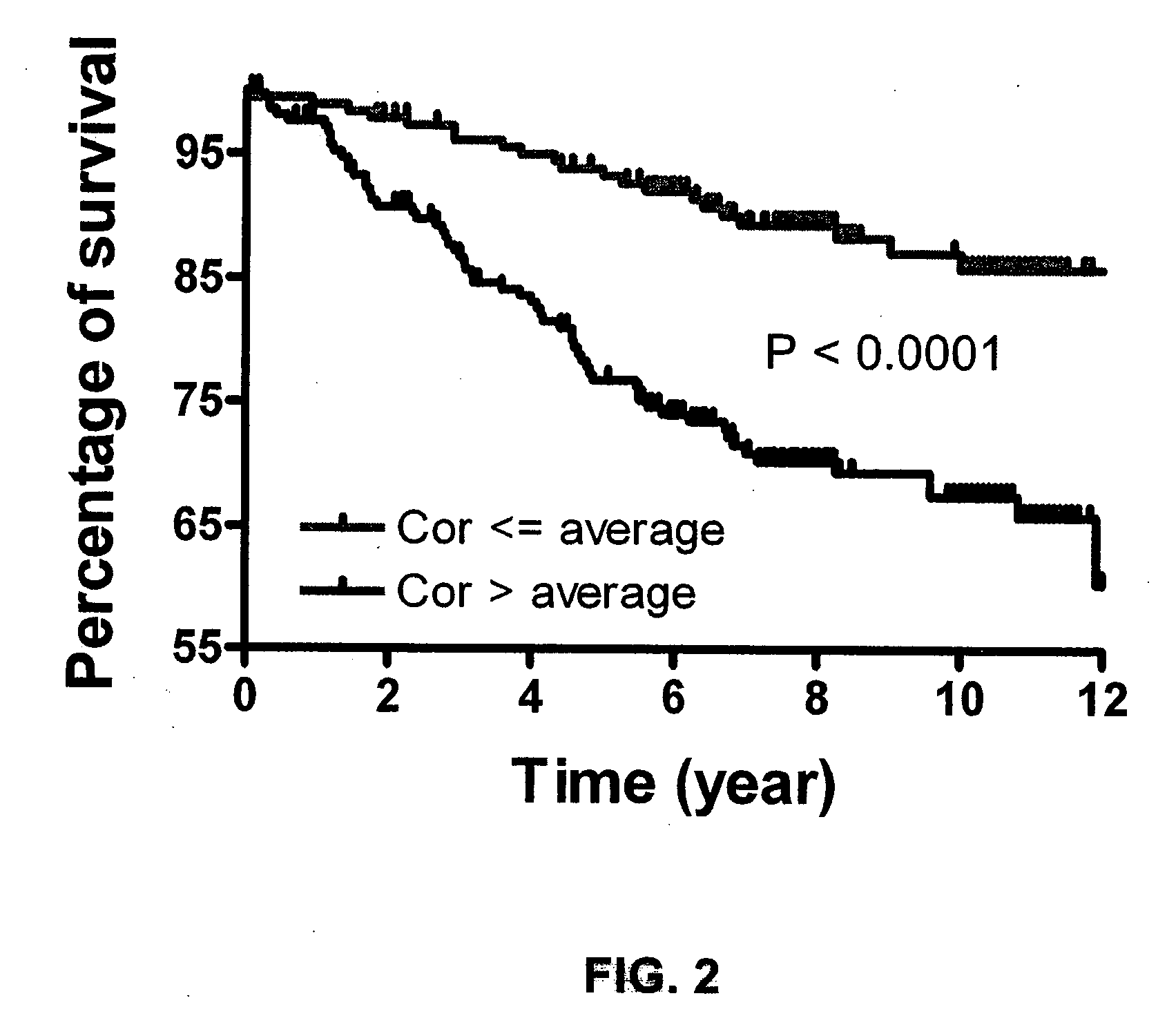
[0203]This example describes the identification of a cancer stem cell gene signature useful to diagnose, predict clinical outcome of a cancer including metastasis and overall survival, as well as for use in clinical trials for achieving stratification of patients and for testing of a potential therapeutic for treating cancer and for guiding cancer therapy. The cancers for which a cancer stem cell gene signature can be used to predict clinical outcome include for example, breast cancer, colon and rectum cancer, pancreas, lung and bronchus, urinary bladder, kidney, head and neck cancer and additional cancers: as set forth in more detail above. Furthermore, use of these cancer stem cell gene signatures to classify tumor samples into low and high risk for the purpose of prognosis and therapy selection is provided.
Cancer Stem Cell Gene Signatures 1 & 2 Generated by Expression Level and P Value
[0204]The genome-wi
PUM
| Property | Measurement | Unit |
|---|---|---|
| Current | aaaaa | aaaaa |
| Current | aaaaa | aaaaa |
| Current | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap