Methods of identifying and modulating pathogen resistance in plants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
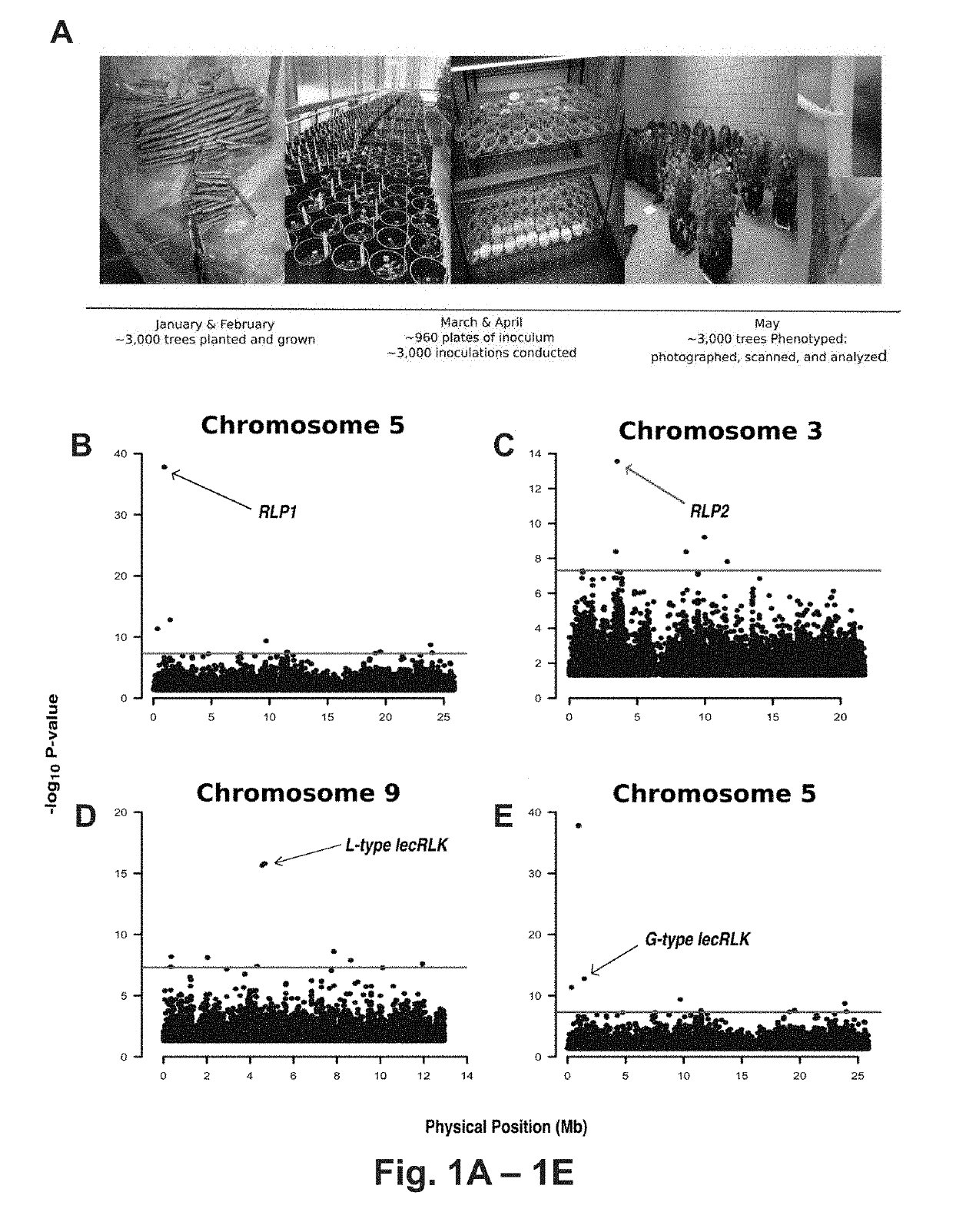
[0120] Discovering S. musiva resistance and susceptibility loci in P. triehoearpa
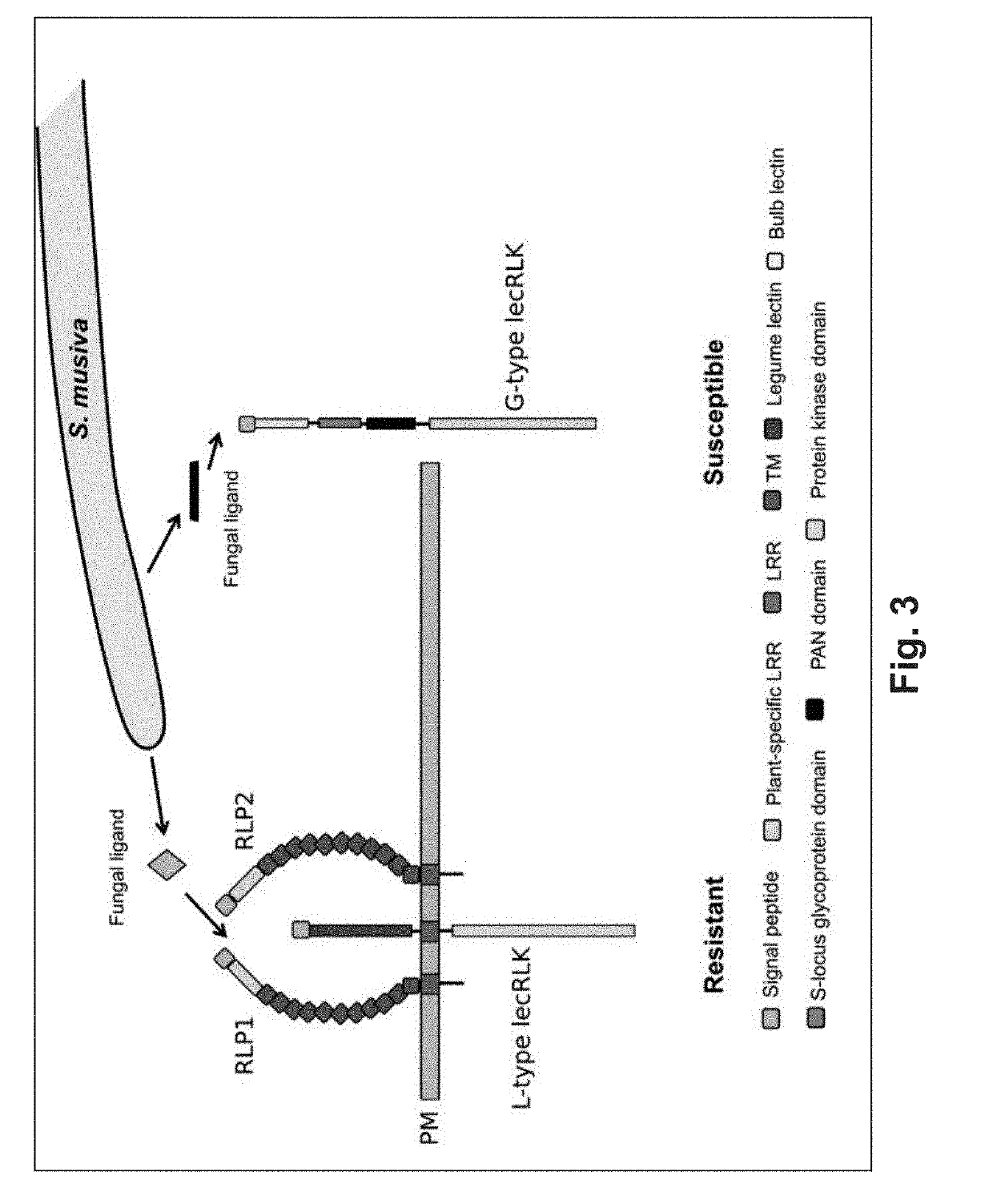
[0121]In a replicated greenhouse experiment 3,404 plants, from a population of 1,081 unrelated trichocarpa genotypes, were characterized for post-inoculation phenotypic responses to S. masiva. Phenotypes were correlated to 8.2 million single nucleotide polymorphisms (SNPs) and insertion / deletions (indels). This process allowed identification of 82 candidate genes encompassing 113 polymorphisms within 5 months of planting the trees (Table 5). Notably, four of the most significant associations were to genes predicted to encode proteins with domains common to pattern recognition receptors (PRRs), including two paralogous leucine-rich receptor-like proteins (RLPs) [Potri.005G012100, p-value=1.56E-38; Potri.003G028200, p-value=2.78E-14], an L-type lectin receptor-like kinase (L-type lecRLK) [Patri.009G036300, p-value=2.115E-16] and a G-type lectin receptor-like kinase (G-type lecRLK) [Potri.005G018000, p-val
example 2
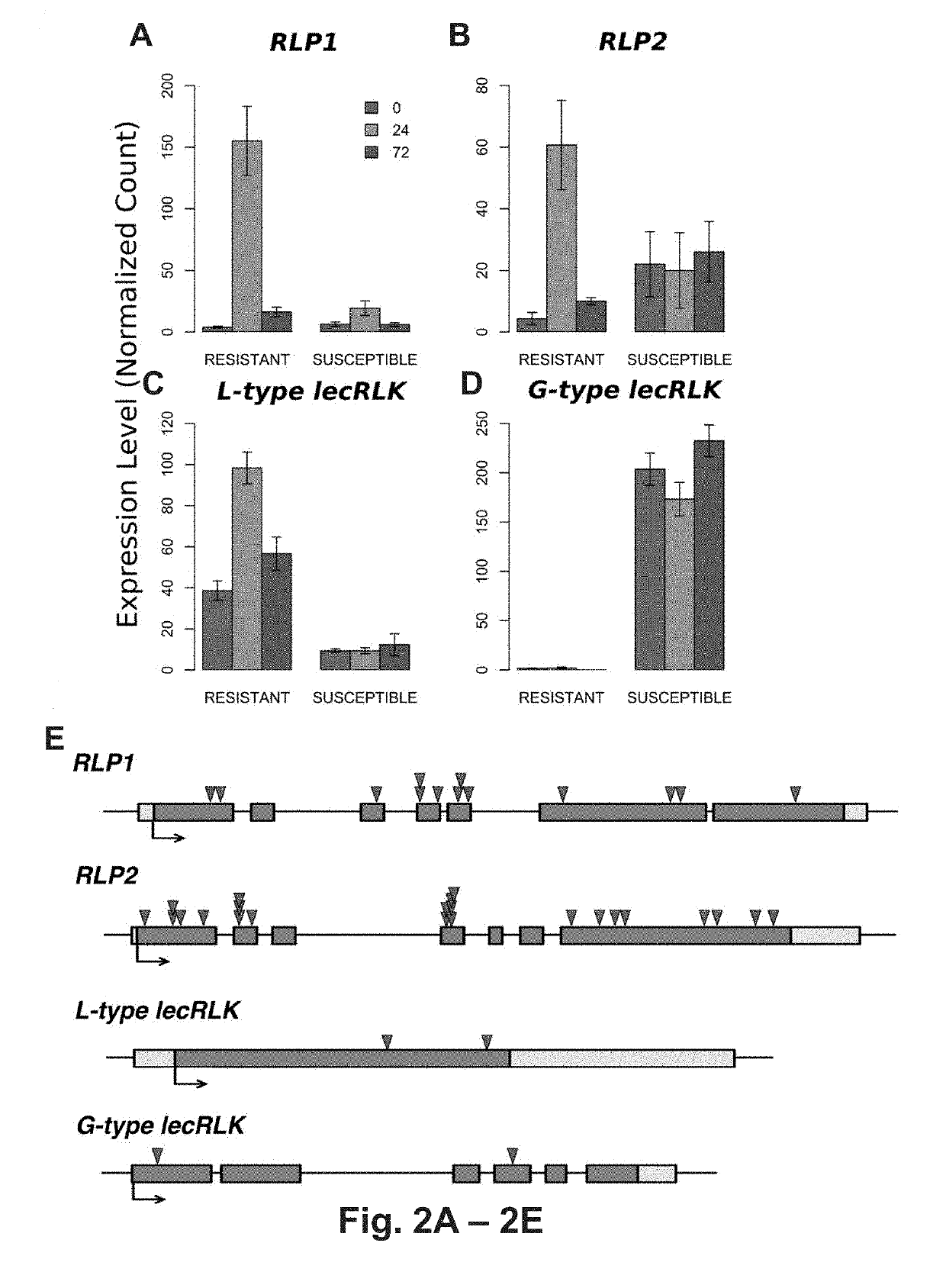
[0123] Transcriptome analysis of resistant and susceptible genotypes
[0124]Transcriptome changes of resistant (BESC-22) and susceptible (BESC-801) genotypes were compared at 0-, 24-, and 72-h post-inoculation (hpi) with S. musiva. Transcriptional changes within (different time points) and between genotypes (same time points) were analyzed. In total 4,872 genes were differentially expressed between the 0- and 72-hpi in the resistant compared to 79 in the susceptible genotype. PFAM domain-enrichment analysis revealed major protein families associated with innate immunity responses, with >2X up-regulation in the resistant genotype and no response in the susceptible genotype. Interestingly, these results are inconsistent with previous observations on co-evolved pathosystems, which suggested that resistant and susceptible responses share similar sets of differentially expressed genes that vary only in timing and amplitude of expression (Chen, W. et al., Plant J., 46, 794-804 (2000).
[0125]A s
example 3
[0126] Population-wide Mutation Analysis of the Susceptibility and Resistance Loci
[0127]To correlate the predicted function of these loci within the P. trichocarpa population with susceptibility and resistance to the fungal pathogen the population-wide occurrence of mutations were examined using a SnpEff analysis (Cingolani P et al., Fly (Austin), 6: 80-92 (2012)). This revealed extensive occurrences of high-impact (deleterious) mutations (early translation termination, frame-shift, and changes in splice-site acceptor, and / or splice-site donor sequences) in the putative resistance-associated RLP-encoding loci (FIG. 2E). In contrast, the putative susceptibility G-type lecRLK locus was highly conserved across the population (FIG. 2E). Only two high-impact mutations were found in 1.5% and 8.0% of the population, respectively. The first is a premature stop codon at position 1441171 bp (G>A) on chromosome 9, that is predicted to truncating the protein to 5% of its length. The second is a
PUM
| Property | Measurement | Unit |
|---|---|---|
| Time | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap