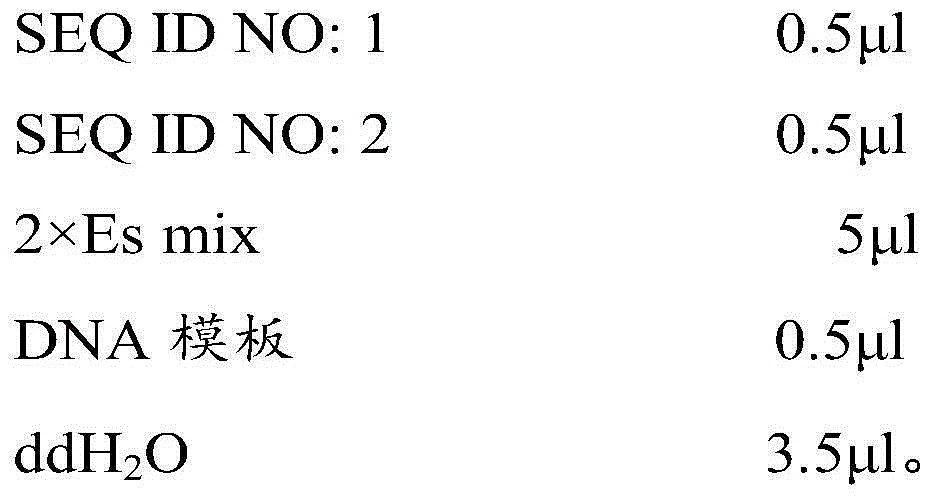Molecular identification primers and method for chicken fast/slow feathering phenotype
A molecular identification, fast and slow technology, applied in the field of molecular genetics, can solve the problems of unclear identification standards, limited identification time, and long time for establishment of male and female matching lines. Good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Example Embodiment
[0040] Example 1
[0041] A molecular identification method for chicken fast and slow feather phenotype, comprising the following steps:
[0042] 1) Extraction of template DNA: usually extracted from chicken blood, of course, it can also be extracted from other tissues;
[0043] 2), with primers shown in SEQ ID NO: 1 and SEQ ID NO: 2, take 1) the extracted DNA as template to carry out PCR amplification;
[0044] 3), carry out agarose gel electrophoresis with the product obtained by PCR amplification in 2);
[0045] 4), analyze the electrophoresis results in 3).
Example Embodiment
[0046] Example 2
[0047] 1. Primer preparation
[0048] Primer synthesis was performed by Sangon Bioengineering (Shanghai) Co., Ltd., including:
[0049] Forward primer 1305s: 5'CCTCCTTGCCAAACCTTA3';
[0050] Reverse primer 1305a: 5'ATCAGCCAGATCCGTCAG3'.
[0051] 2. The steps of the kit to extract chicken blood clot DNA:
[0052] The ClotBloodDNAKit of Kangwei Century Biotechnology Co., Ltd. was selected for extraction, product number CW0545.
[0053](1) Weigh 0.1-0.2 g of coagulated blood clot, place it in a centrifuge tube, add 550 μl BufferGTL, and homogenize with an electric homogenizer. In order to fully crush the blood clot, small magnetic beads can be added for mechanical crushing.
[0054] (2) Add 20 μl of proteinase K and invert the sample upside down to fully mix the sample. Incubate overnight at 56°C in a shaker until all particles are completely dissolved.
[0055] (3) Add 200 μl BufferPP, vortex for 20 seconds, and incubate on ice for 3-5 minutes. Centrifuge
Example Embodiment
[0079] Example 3
[0080] 1. Primer design and preparation
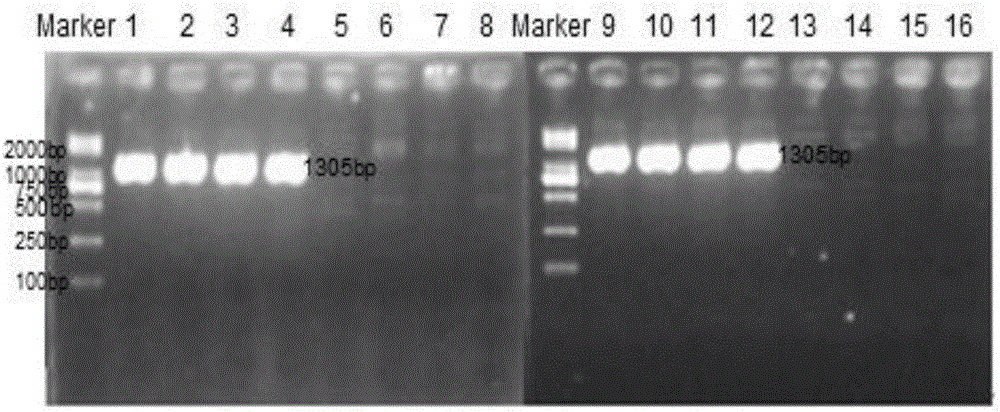
[0081] Elferink et al. (2008) found that the slow feather K allele that controls feather speed is composed of tandem repeats with a total length of 176324bp, caused by partial duplication of two genes: PRLR gene and SPEF2 gene encoding sperm flagellin, including Exon 1-exon 11 and 558bp exon 12 of the PRLR gene and exon 1-exon 5 of the SPEF2 gene were identified; and primers were designed to amplify a 78bp long The breakpoint connects the sequence, but the fast feather chicken cannot amplify this sequence, so it can be used as a molecular detection method to distinguish fast and slow feathers. On the basis of the research results, primers were designed to amplify a breakpoint junction sequence containing 78bp and a length of 1305bp, which can only be amplified by slow-feathered chickens, but not by fast-feathered chickens, so that chickens can be distinguished The fast and slow feather type. The fragment is easy to a
PUM
| Property | Measurement | Unit |
|---|---|---|
| Total length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap