Identification and application of pig CD4 gene function mutation site molecular breeding marker
A site and application technology, applied in the identification and application field of molecular breeding markers of porcine CD4 gene functional mutation sites, can solve the problems of unreported porcine CD4 gene functional mutations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
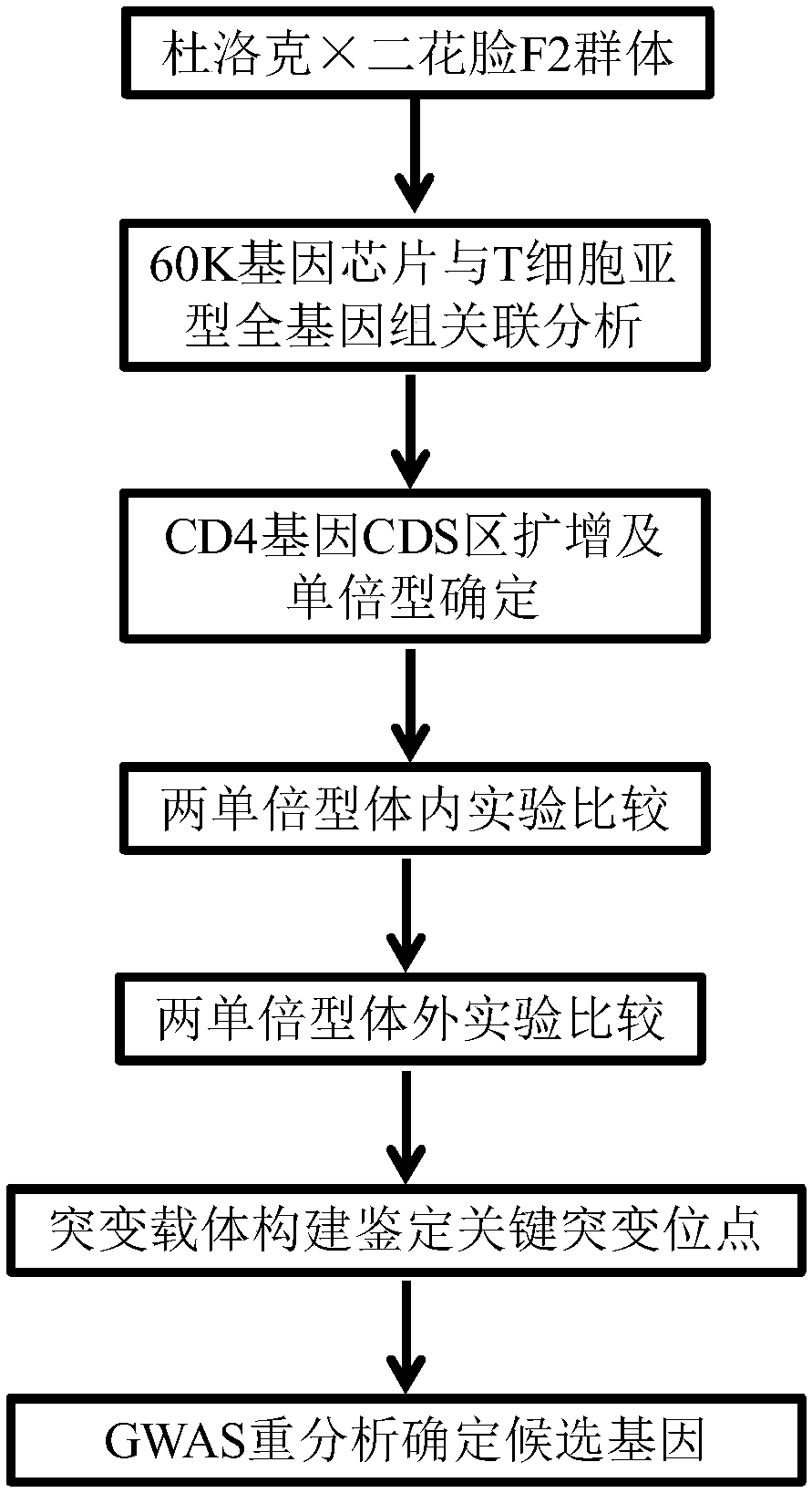
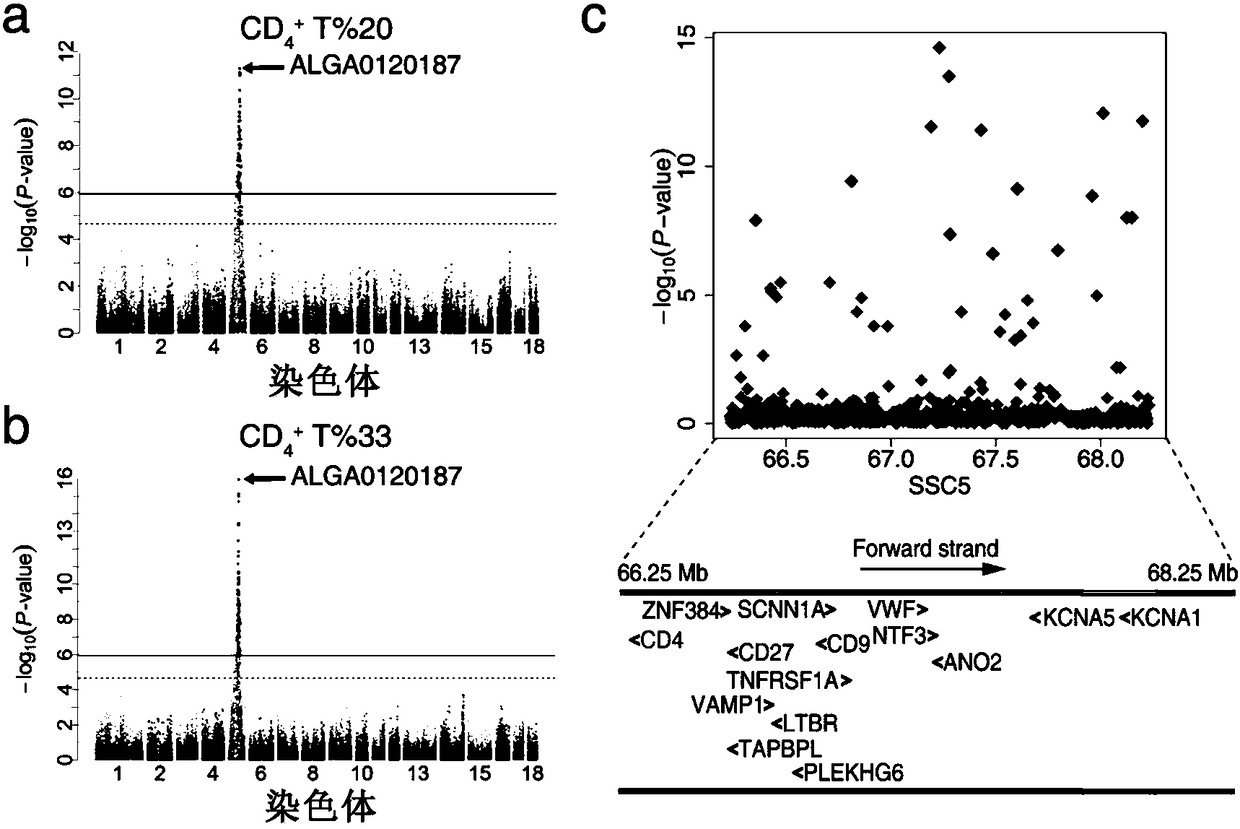
[0124] Genome-wide association analysis (GWAS) of porcine 60K gene chip and T cell subtypes
[0125] The 60K gene chip (Illumina, USA) of the Duroc × Erhualian F2 population was used to analyze the relationship between each SNP and the T cell subtype using a mixed model. The model can be expressed as Y=Xb+Sα+Zμ+e, where Y is the table type vector, b is the estimated value of fixed effect gender and batch, μ is the additive effect of gene (μ-N(0,Gσ α 2 ), G is the genome similarity matrix, σ α 2 is the polygenic additive variance), X and Z are the correlation matrices of b and μ, S is the correlation vector of α, and e is the residual matrix (in the form of N(0,Iσ e 2 ) distribution, where I is the identity matrix, σ e 2 is the residual variance). For specific analysis, the software package in R was used, referring to the research of Aulchenko et al. (Aulchenko et al., 2007a,b), and the thresholds for genome-level significance and suggested significance were set after Bonfe
Embodiment 2
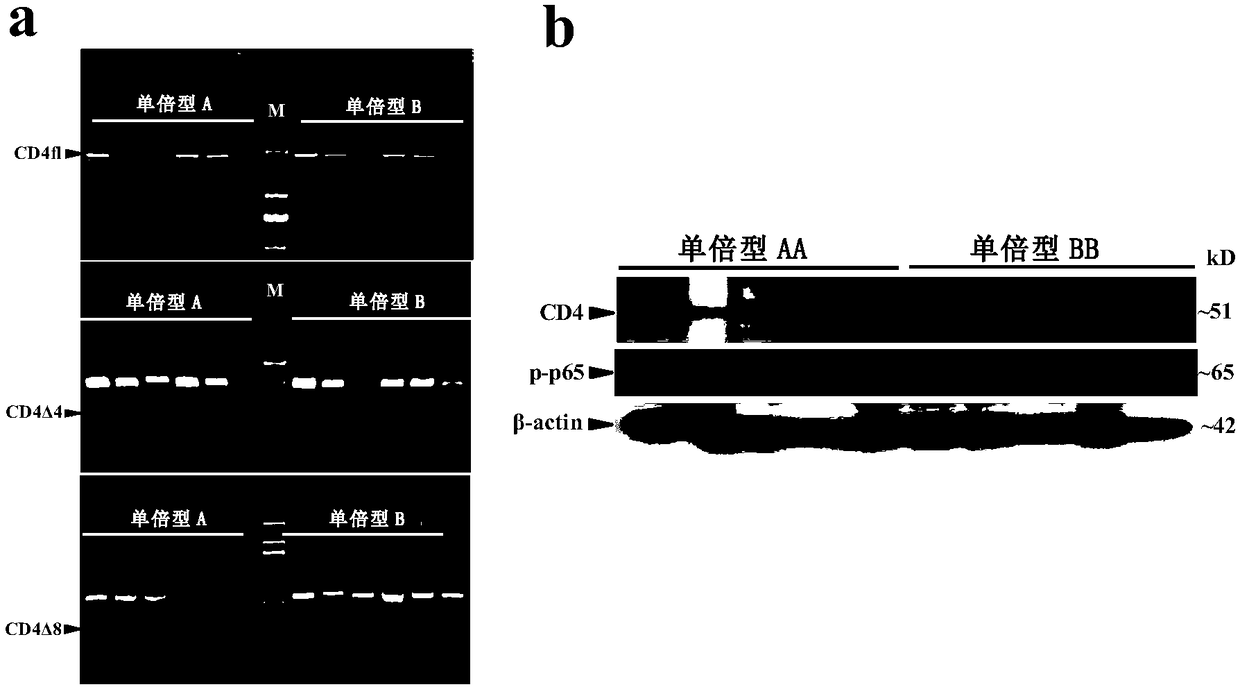
[0127] Determination of two haplotypes of porcine CD4 gene
[0128] The experimental method is as follows:
[0129] (1) Using TRIZOL lysis method to extract total white blood cell RNA from Duroc × Erhualian F2 population
[0130] 1) Take out the white blood cell lysate added with 1ml TRIZOL from -80°C, dissolve it on ice, shake vigorously to lyse the cells;
[0131] 2) Add 0.2ml of chloroform, shake vigorously for 15s, let stand at room temperature for 2-3min, RNA is dissolved in the supernatant water sample layer, DNA and protein are dissolved in the organic layer below;
[0132] 3) Place the centrifuge tube in a refrigerated centrifuge, centrifuge at 12000rpm at 4°C for 15min, carefully absorb the water sample layer and transfer it to a new 1.5ml centrifuge tube, add an equal volume of isopropanol, mix well and let stand at room temperature for 15min to precipitate total RNA;
[0133] 4) Place the centrifuge tube in a refrigerated centrifuge, centrifuge at 12,000 rpm at 4°C
Embodiment 3
[0169]Mutation vector construction and identification of key mutation sites
[0170] The experimental method is as follows:
[0171] (1) Mutation vector construction
[0172] Using the haplotype A and haplotype B overexpression vectors as templates, design mutant vector primers with primer sequences such as SEQ ID NO: 5-8, and carry out the amplified SNPs region containing type A and the SNPs region containing type B To reassemble the connection, the specific steps are as follows:
[0173] 1) PCR amplification recovery. Using the constructed haplotype A and haplotype B vectors as templates, PCR amplification was performed with primer pairs of SEQ ID NO: 5-8, and the amplification system was as follows (50 μL):
[0174]
[0175] After mixing and centrifuging, perform PCR amplification. The reaction procedure is as follows:
[0176]
[0177] The PCR product was recovered by gel, and the recovered product was detected by electrophoresis.
[0178] 2) Recombination connecti
PUM
| Property | Measurement | Unit |
|---|---|---|
| Aperture | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap