Mycobacterium tuberculosis (MTB) H37Rv coding gene and application thereof
A technology of Mycobacterium tuberculosis and coding genes, which is applied in application, genetic engineering, plant genetic improvement, etc., can solve problems such as gene boundary errors, ribosome translocation, and termination site errors, and achieve the effect of easy comparison
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Example Embodiment
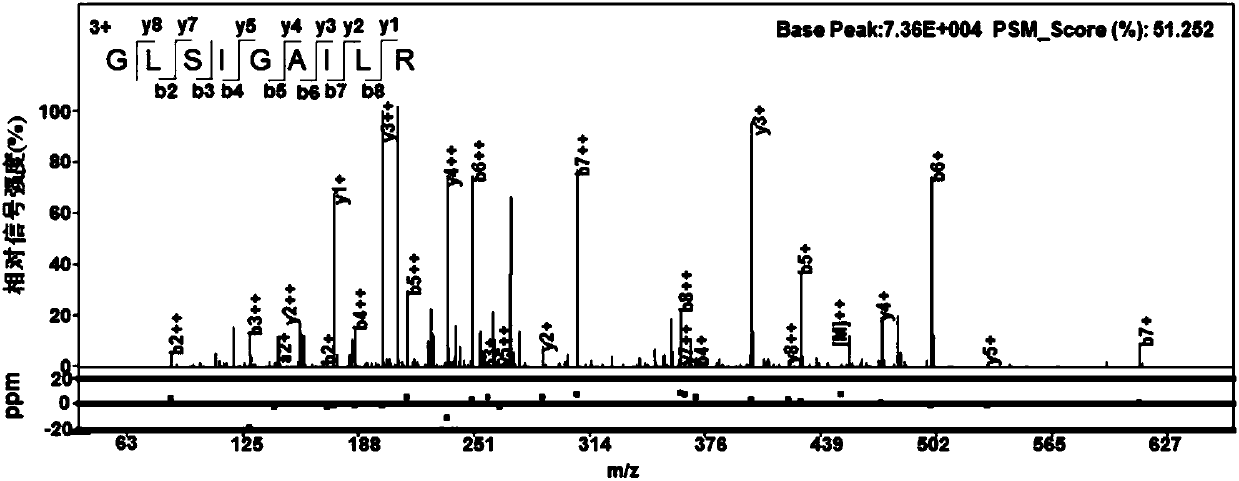
[0033] Example 1: Searching for missing-annotated coding genes in the genome of H37Rv strain
[0034] 1.1 High-coverage proteomic verification of the H37Rv strain genome
[0035] Using high-coverage proteomics technology, the H37Rv strain was used to study the deep proteomic coverage. Based on the Tuberculosis (20160307) database, the pFind 3 engine was used to validate the annotated coding genes of its genome. In order to discover new protein coding regions, based on protein genomics technology, we used pAnno software to translate the H37Rv genome-wide (NC_000962.3) file published in NCBI on a six-reading frame database, and used this database to perform new peptides on mass spectrometry data. Identification of segments and new proteins. In order to reduce the false positive rate, we used three filtering methods to estimate the category FDR separately for the annotated peptides and the new peptides in the data filtering process, namely S-FDR, T-FDR I and T-FDRII.
[0036] Through da
Example Embodiment
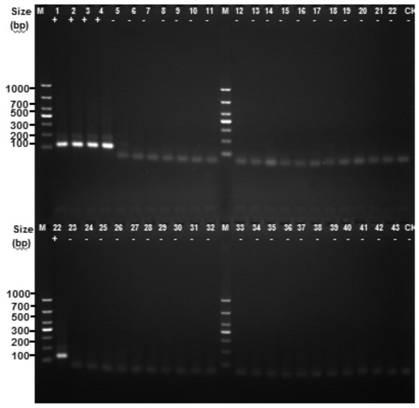
[0051] Example 2: Establishment of a method for identifying MTBC complex
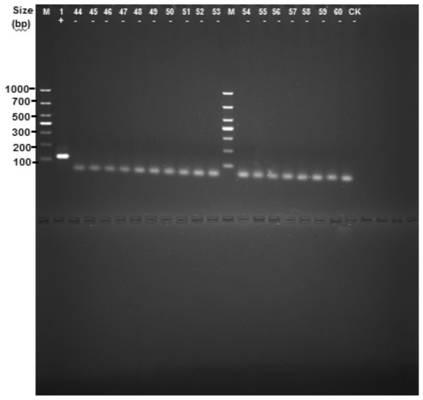
[0052] (1) Design primers:
[0053] Based on the CDS sequence of the Rv1796c(-|2034439-2034570|) gene shown in SEQ ID NO.1, PCR primers were designed using Oligo7.0, the primer sequences are as follows:
[0054] F: 5'-ATGGAGATTATCTCGACACCGGGGG-3' (SEQ ID NO. 4);
[0055] R: 5'-GCAAGATAGCCCCGATCGACAACCC-3' (SEQ ID NO.5)
[0056] The positional relationship between the above primers and the Rv1796c(-|2034439-2034570|) gene is shown below, and the corresponding positions of the primers are marked with a single underline.
[0057] ATGGAGATTATCTCGACACCGGGGG CGATCCCACTGAACGCGTCCGGCGGTGGTCCCAGCGCTGGCGATGGCGCACTCGGGTGCGGATTACGC GGGTTGTCGATCGGGGCTATCTTGC GGCCCCCGGAGTAG (SEQ ID NO.1)
[0058] (2) Extract the total DNA of the tested strains including M. tuberculosis H37Rv, 40 standard strains of Mycobacterium are deposited by the Chinese Medical Bacterial Culture Collection and Management Center (CMCC), and the remaining 16 s
PUM
| Property | Measurement | Unit |
|---|---|---|
| Theoretical molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap