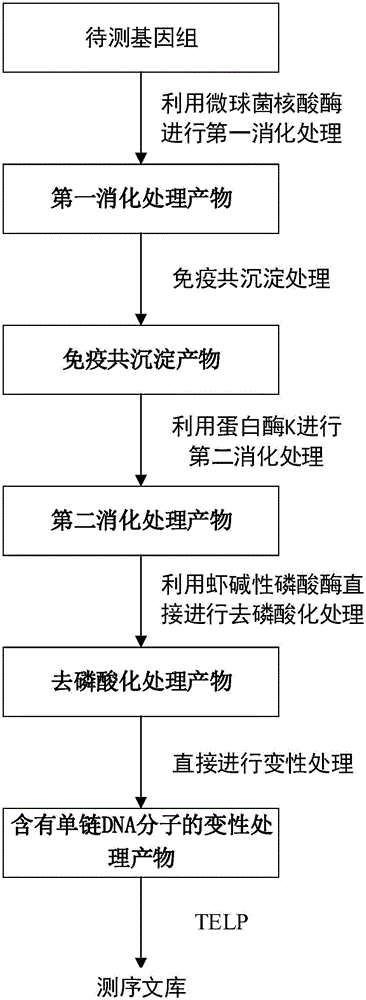
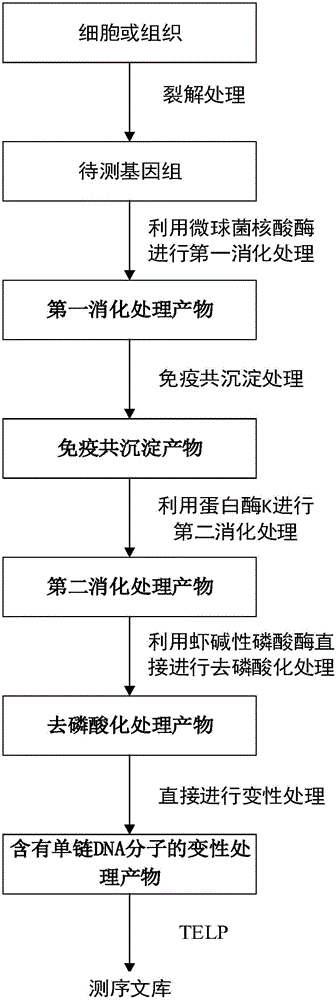
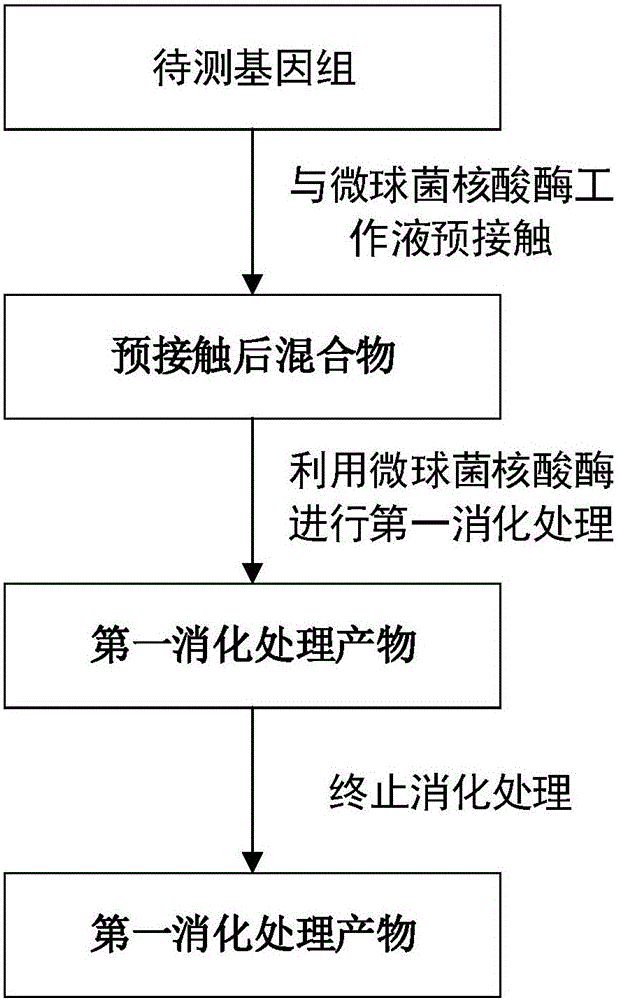
Method for constructing DNA sequencing library of genome under detection and application of method
A DNA sequencing and sequencing library technology, applied in the biological field, can solve the problems of limited antibody quality and inability to obtain effective histone modification information, so as to avoid local damage and improve recognition efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0088] Example 1 Construction of DNA sequencing library of cell genome
[0089] 1.1 Reagent preparation
[0090] Lysis buffer
[0091] 0.5% NP-40
[0092] 0.5%Tween
[0093] 0.1% SDS
[0094] MNase working buffer (MNase working buffer)
[0095] ·100mM Tris-HClph8.0
[0096] 2mM CaCl2
[0097] MNase Dilute buffer (MNaseDilutebuffer)
[0098] ·50mM Tris‐HCl, pH8.0
[0099] 1mM CaCl2
[0100] 0.2% Triton X-100
[0101] 10xMNase stop buffer (10xMNaseStopbuffer)
[0102] 110mM Tris-HCl pH8.0
[0103] 55mM EDTA
[0104] 2xRIPA buffer2xRIPAbuffer(onice)
[0105] 280mM NaCl
[0106] 1% Triton X‐100
[0107] 0.1% SDS
[0108] 0.2% Na‐Deoxycholate
[0109] 5mMEGTA
[0110] RIPA buffer RIPAbuffer
[0111] ·10mM Tris pH8.0
[0112] 1 mM EDTA
[0113] 140mM NaCl
[0114] 1% Triton X‐100
[0115] 0.1% SDS
[0116] 0.1% Na‐Deoxycholate
[0117] Lithium chloride buffer (LiClwashbuffer)
[0118] 250mMLiCl
[0119] ·10mM Tris pH8.0
[0120] 1 mM EDTA
[0121] 0.5% NP-4
Embodiment 2
[0137] Example 2 Screening of dephosphorylation enzymes
[0138] In order to obtain the best dephosphorylated end repair results and ensure the final DNA yield, the inventors screened the most suitable dephosphorylation treatment enzyme from the three combinations of T4PNK, T4DNA polymerase, Klenow or T4PNK or rSAP. The specific experimental process is as follows Said:
[0139] The mouse embryonic stem cells grown on the six-well cell culture plate were taken out of the incubator, the medium was sucked off, and the medium was gently washed with 1ml of PBS to wash off the residual medium. Then add 0.5ml of trypsin with a concentration of 0.05%, digest at 37° for three minutes, and stop the digestion reaction with 1ml of fresh medium. In order to make the cells completely become a single suspended cell state, pipette repeatedly with a 1ml large pipette until no cell clumps are visible to the naked eye. Then centrifuge at a low speed of 2000rmp for 5 minutes. After the supernatan
Embodiment 3
[0159] In order to verify the effectiveness of the library construction method proposed in this application, the inventors adopted the most stringent testing method: through co-immunoprecipitation DNA sequencing technology based on a large number (1x10e7) of mouse embryonic stem cells (1x10e7) in the ENCODE database (ChIP-sequence) results were compared to see the correlation between the two.
[0160] Specifically, the inventor compared the map data obtained by the inventor with the ENCODE map data, and loaded it into UCSCbrowser. Then, by observing the peak distribution of H3K4me3 and combining its positional relationship with the gene promoter, the inventor judged the Whether the method for building the database proposed by the invention is successful.
[0161] The result is as Figure 16 As shown, it can be seen from the results (genomic position: chr6:51,271,690-52,394,589), whether it is the result (ENCODE) obtained from millions of cells or the ChIP-sequence result of the
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap