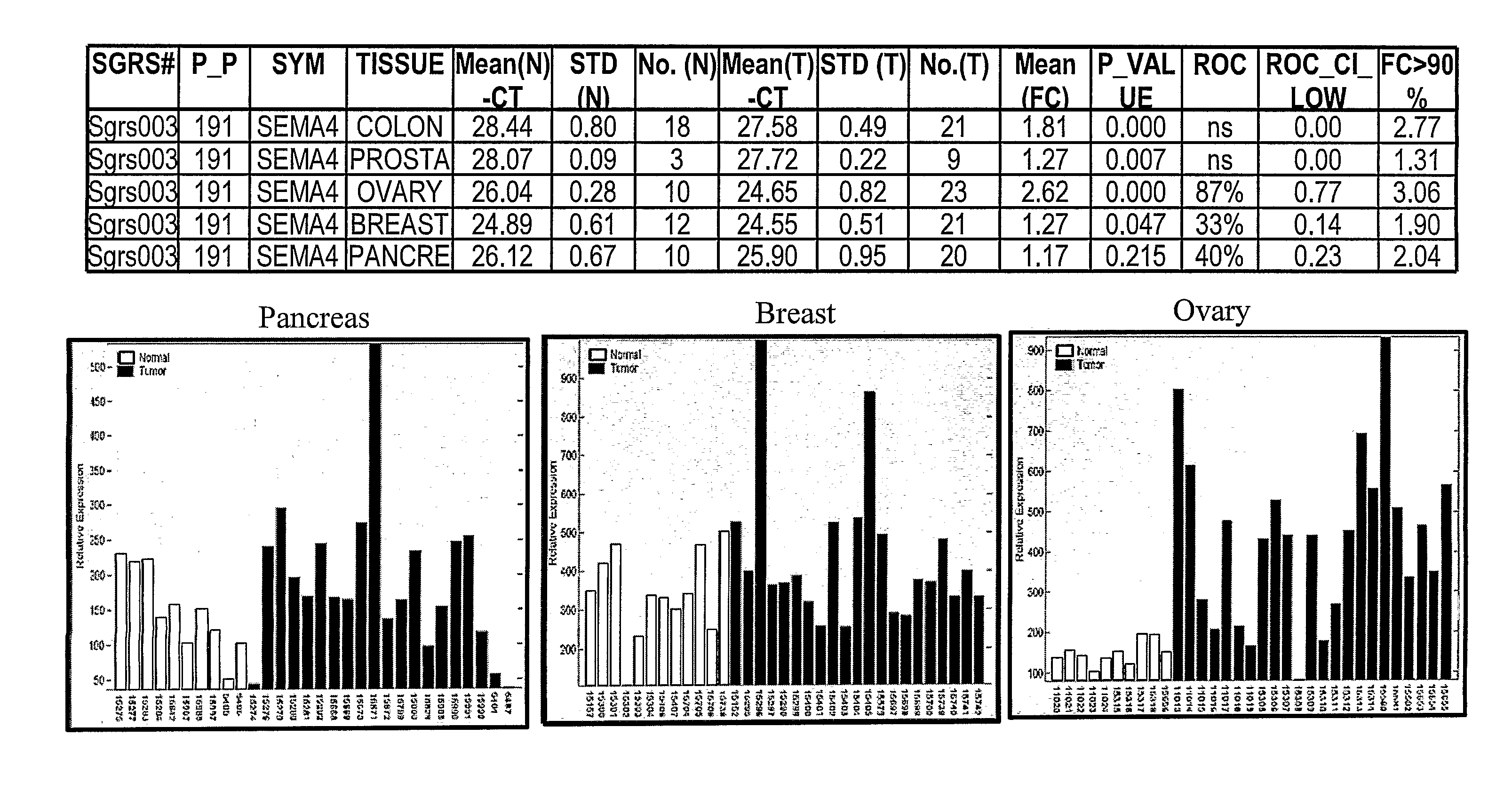
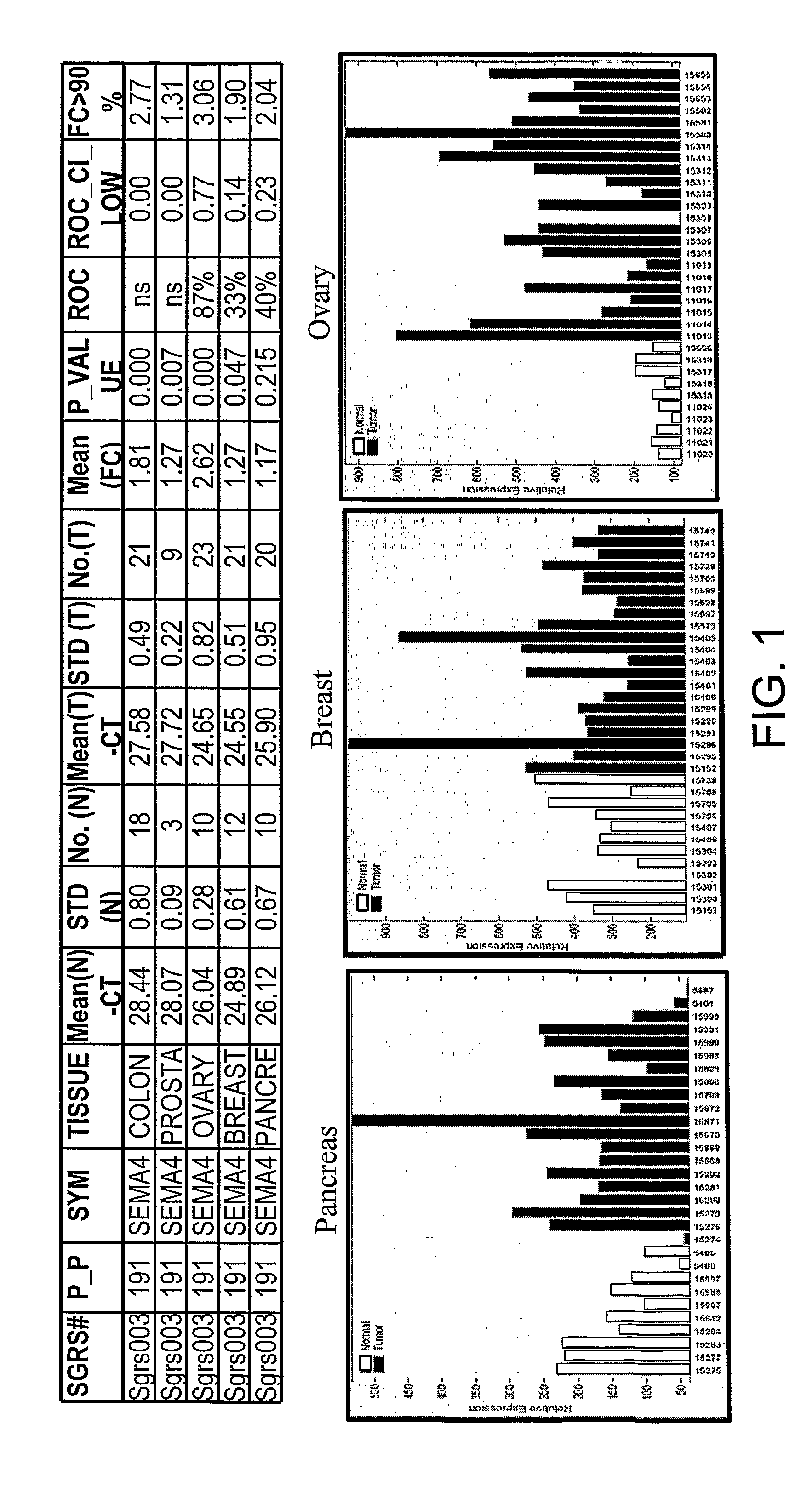
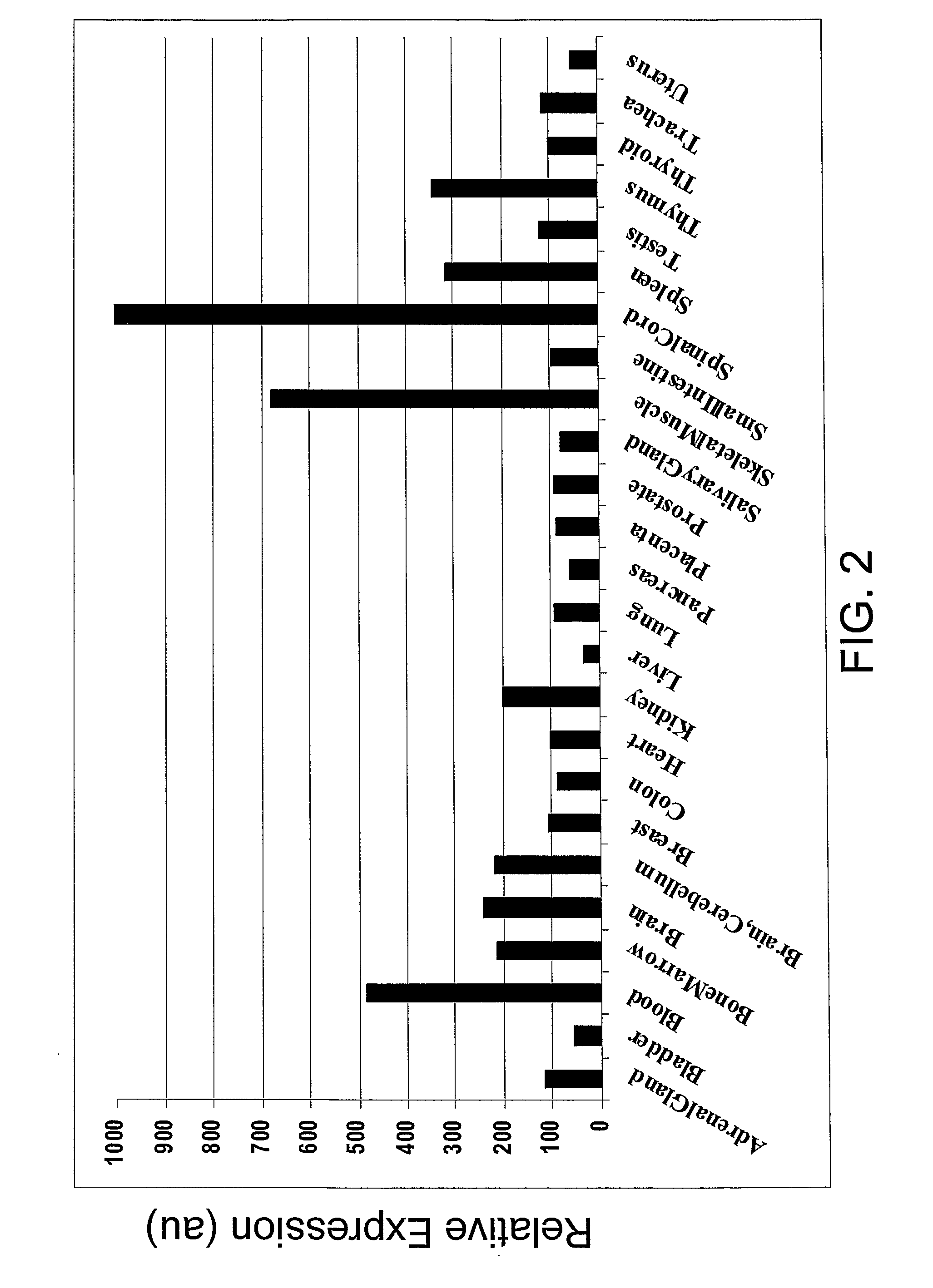
SEMA4D in Cancer Diagnosis, Detection and Treatment
a cancer diagnosis and detection technology, applied in the field of sema4d in cancer diagnosis, detection and treatment, can solve problems such as truncation or unstable transcripts/protein products
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Insertion Site Analysis Following Tumor Induction in Mice
[0379]Tumors were induced in mice using either mouse mammary tumor virus (MMTV) or murine leukemia virus (MLV). MMTV causes mammary adenocarcinomas and MLV causes a variety of different hematopoetic malignancies (primarily T- or B-cell lymphomas).
[0380]Three routes of infection were used: (1) injection of neonates with purified virus preparations, (2) infection by milk-borne virus during nursing, and (3) genetic transmission of pathogenic proviruses via the germ-line (Akvr1 and / or Mtv2). The type of malignancy present in each affected mouse was determined by histological analysis of H&E-stained thin sections of formalin-fixed, paraffin-embedded biopsy samples. Host DNA sequences flanking all clonally-integrated proviruses in each tumor were recovered by nested anchored-PCR using two virus-specific primers and two primers specific for a 40 bp double stranded DNA anchor ligated to restriction enzyme digested tumor DNA. Amplified
example 2
Analysis of Quantitative RT-PCR Comparative CT Method
[0385]The RT-PCR analysis was divided into 4 major steps: 1) RNA purification from primary normal and tumor tissues; 2) Generation of first strand cDNA from the purified tissue RNA for Real Time Quantitative PCR; 3) Setup RT-PCR for gene expression using ABI PRISM 7900HT Sequence Detection System tailored for 384-well reactions; 4) Analyze RT-PCR data by statistical methods to identify genes differentially expressed (up-regulated) in cancer. These steps are set out in more detail below.
A) RNA purification from primary normal and tumor tissues
[0386]This was performed using Qiagen RNeasy mini Kit CAT#74106. Tissue chucks typically yielded approximately 30 μg of RNA resulting in a final concentration of approximately 200 ng / gl if 1504 μl of elution buffer was used.
[0387]After RNA was extracted using Qiagen's protocol, Ribogreen quantitation reagents from Molecular Probes was used to determine yield and concentration of RNA according t
example 3
Detection of Cancer-Associated-Sequences in Human Cancer Cells and Tissues
[0411]DNA from prostate and breast cancer tissues and other human cancer tissues, human colon, normal human tissues including non-cancerous prostate, and from other human cell lines are extracted following the procedure of Delli Bovi et al. (1986, Cancer Res. 46:6333-6338). The DNA is resuspended in a solution containing 0.05 M Tris HCl buffer, pH 7.8, and 0.1 mM EDTA, and the amount of DNA recovered is determined by microfluorometry using Hoechst 33258 dye. Cesarone, C. et al., Anal Biochem 100:188-197 (1979).
[0412]Polymerase chain reaction (PCR) is performed using Taq polymerase following the conditions recommended by the manufacturer (Perkin Elmer Cetus) with regard to buffer, Mg2+, and nucleotide concentrations. Thermocycling is performed in a DNA cycler by denaturation at 94° C. for 3 min. followed by either 35 or 50 cycles of 94° C. for 1.5 min., 50° C. for 2 min. and 72° C. for 3 min. The ability of the PC
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap