SNP (single nucleotide polymorphism) marker related to Erhualian sow litter traits and detection method and application thereof
A Erhualian sow and marker technology, which is applied in the field of molecular biology, can solve the problems of not being able to determine the main effect gene and its key mutation sites, and achieve important economic benefits and social value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] 1. Source of experimental animals
[0029] Jiaoxi Erhualian pig professional cooperative in Changzhou, Jiangsu.
[0030] Calculate the breeding value of 177 Erhualian sows, and the calculation model is Y (total litter size) = parity (parity) + farm (field) + year (annual) + season (season) + age (age of sow farrowing )+sire (with boars)+permanent effect+additive effect (permanent effect of sows)+e (residual),
[0031] These include fixed effects - parity, farm / year / season sow farrowing, covariates - sow age at farrowing, random
[0032] Effects - mated boars, permanent effects - sows, individual additive genetic values. Select 18 individuals whose breeding value ranks first or last and whose litter record is more than 3 litters.
[0033] 2. Genomic DNA extraction
[0034] Ear tissue samples from 36 sows were collected, placed in centrifuge tubes filled with 70% alcohol, and stored in a -20°C refrigerator for later use.
[0035] Using the traditional phenol / chlor
Embodiment 2
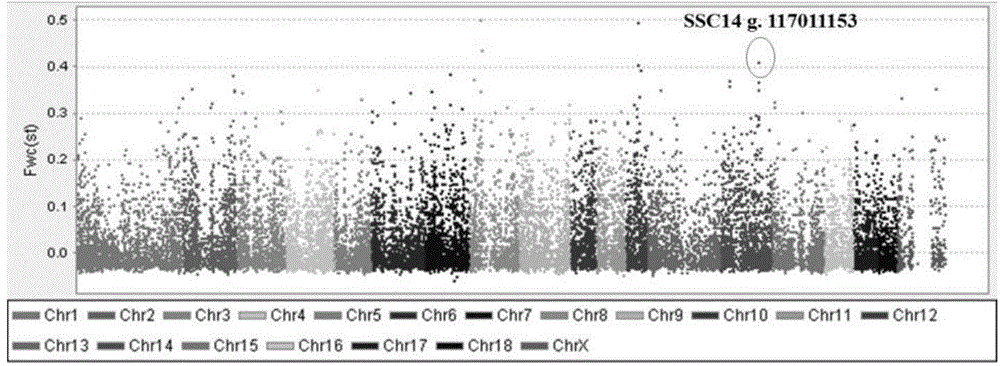
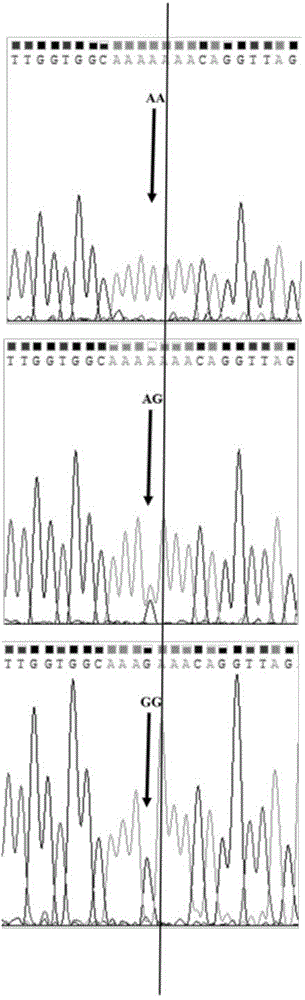
[0059] This example is the verification of the SNP site g.117011153A / G obtained in Example 1 in the Erhualian sow population.
[0060] 1. Extraction of Genomic DNA from Erhualian sows
[0061] Ear tissue samples from 131 purebred Erhualian sows with accurate records of total litter size were collected, placed in centrifuge tubes filled with 70% alcohol, and stored in a -20°C refrigerator for later use. Genomic DNA from ear tissue was extracted by the above method, and after quality and concentration detection, the concentration was diluted to 30 ng / μL and stored at -20°C for future use.
[0062] 2. PCR amplification and sequencing of the target fragment
[0063] Using the extracted DNA as a template, carry out PCR amplification according to the designed primers: take 2.5 μL of DNA template, 1.25 μL of primers shown in SEQ ID NO: 2 and SEQ ID NO: 3, 25 μL of PCR Mix reagent, double 20 μL of distilled water; set up the PCR amplification system: pre-denaturation at 96°C for 2
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap