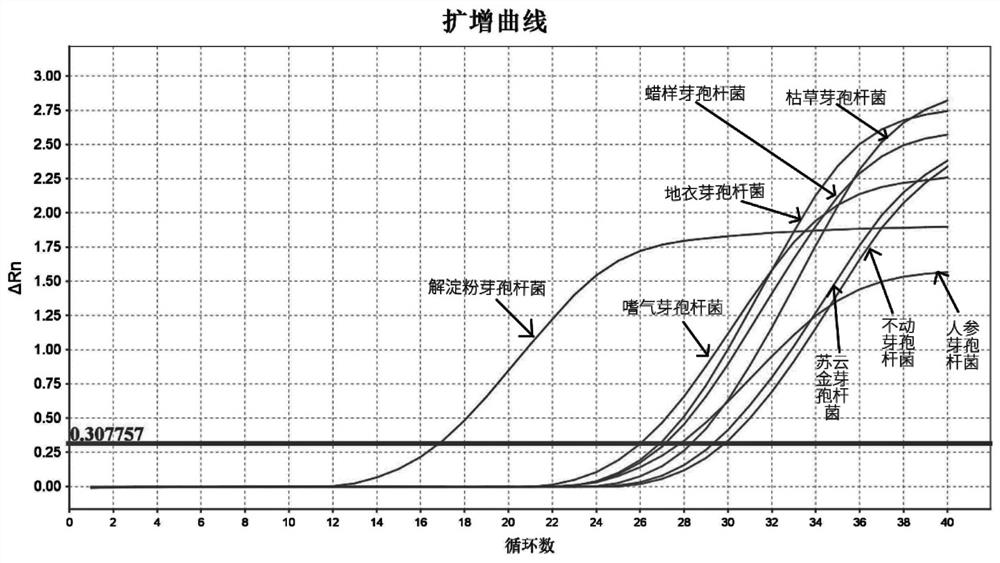
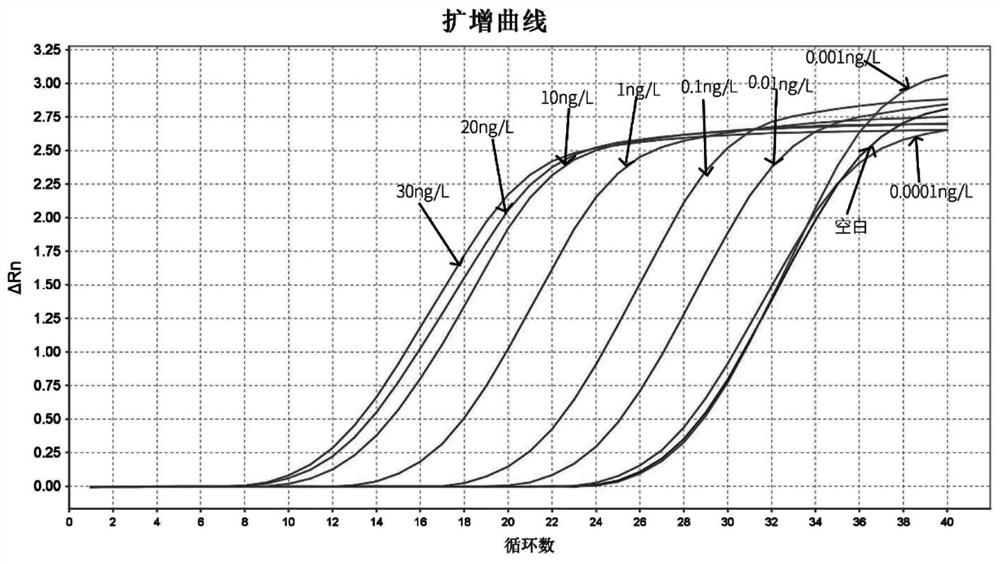
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
42 results about "Nucleotide sequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A nucleic acid sequence is a succession of letters that indicate the order of nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule. By convention, sequences are usually presented from the 5' end to the 3' end. For DNA, the sense strand is used.
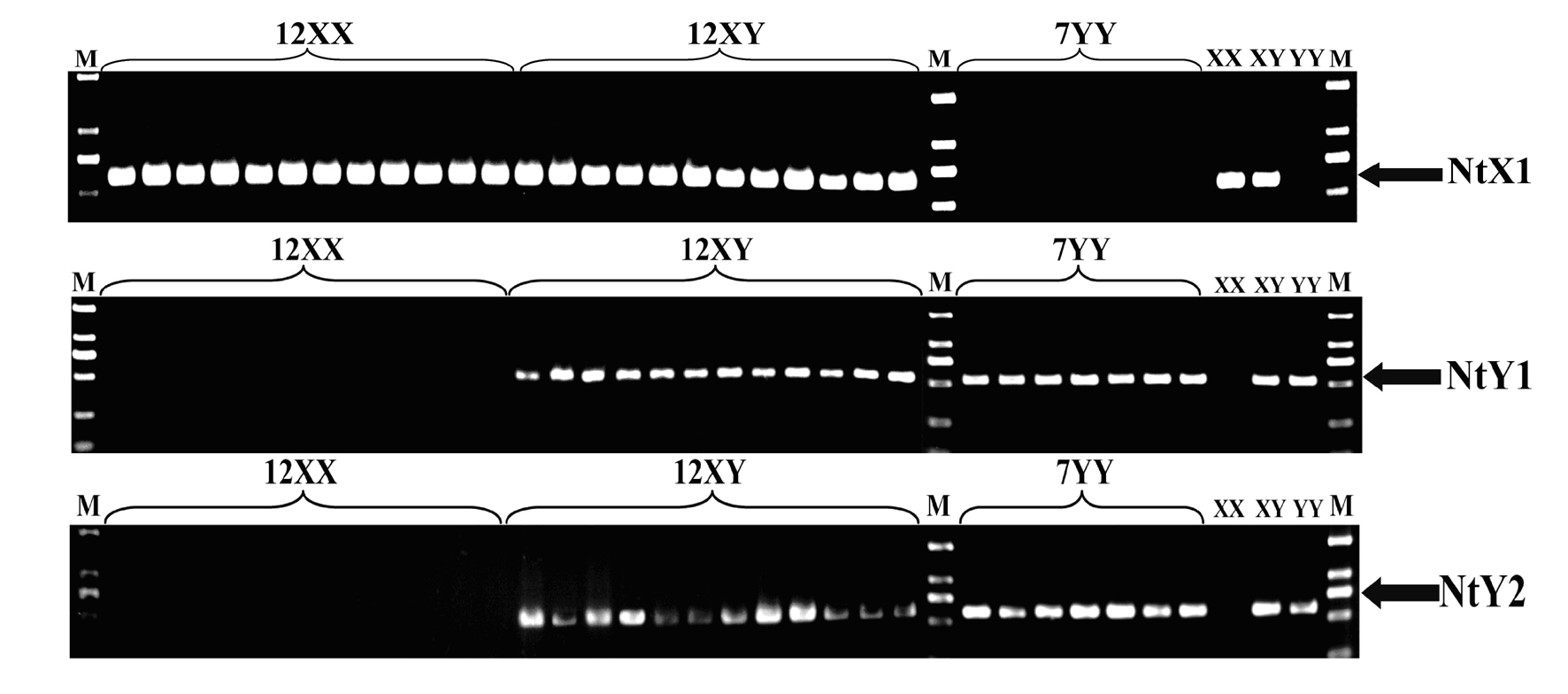
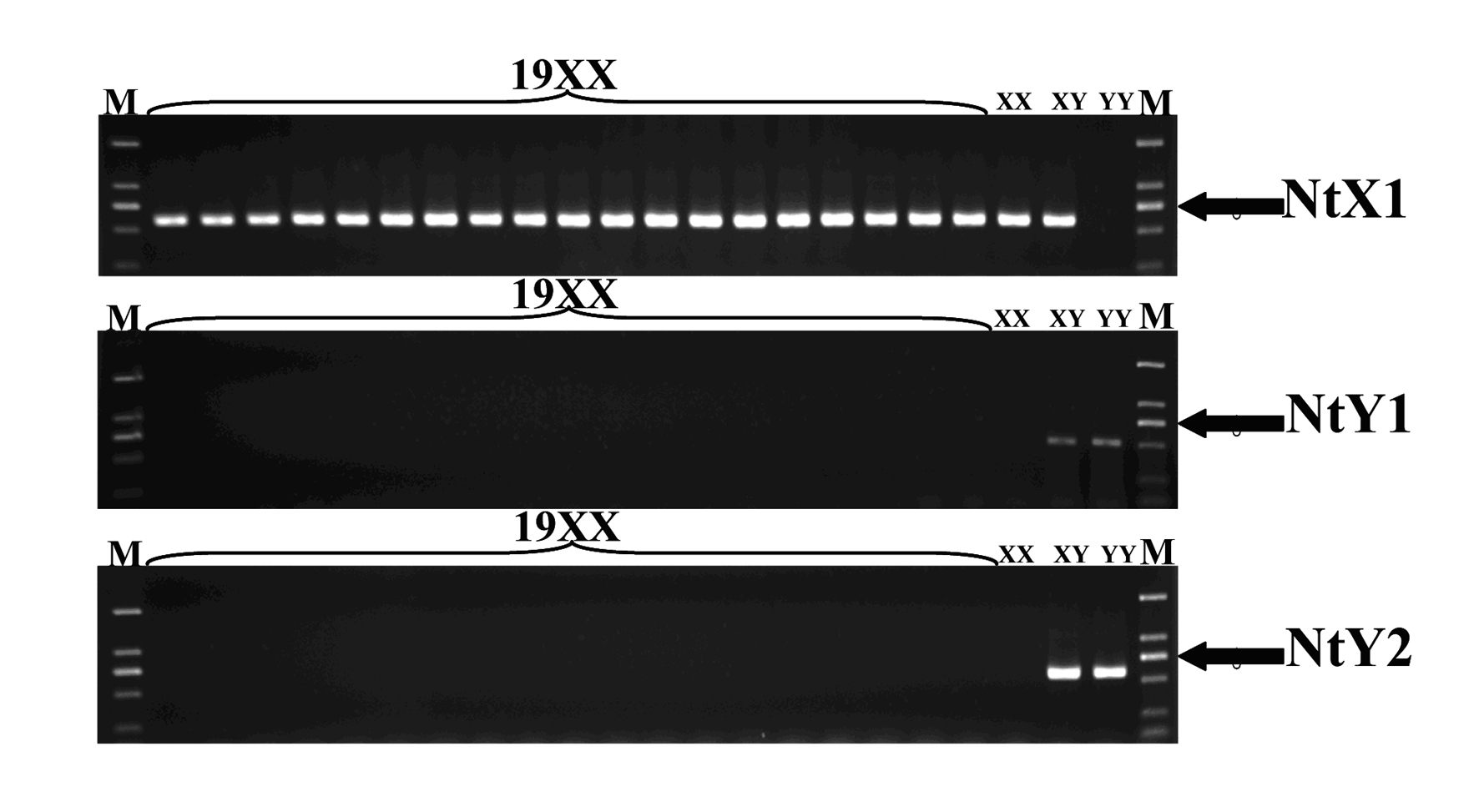
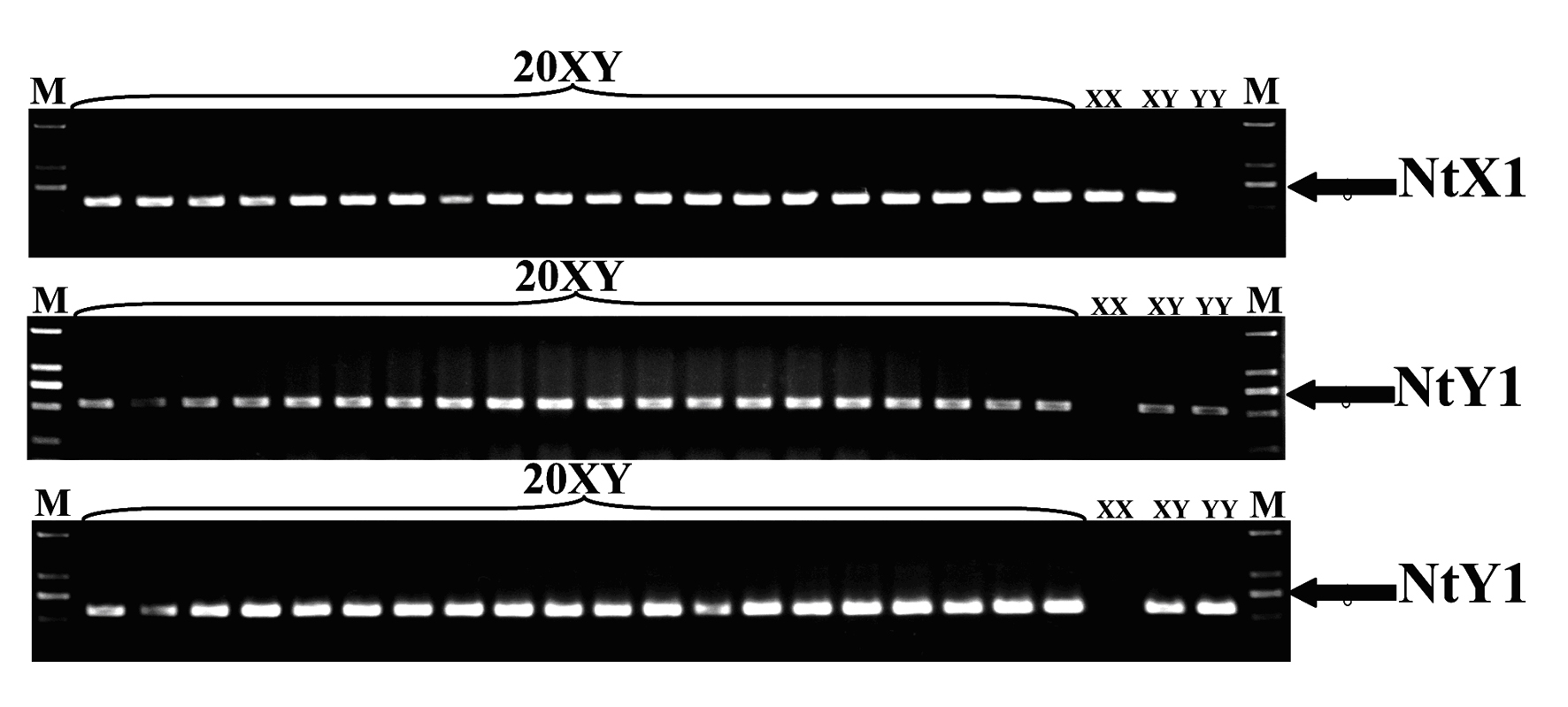
Specific molecular markers of sex chromosomes of Tilapia nilotica and genetic sex identification method
ActiveCN101962641AEconomic divisionQuick distinctionMicrobiological testing/measurementDNA/RNA fragmentationX chromosomeNile tilapia
Owner:SOUTHWEST UNIVERSITY
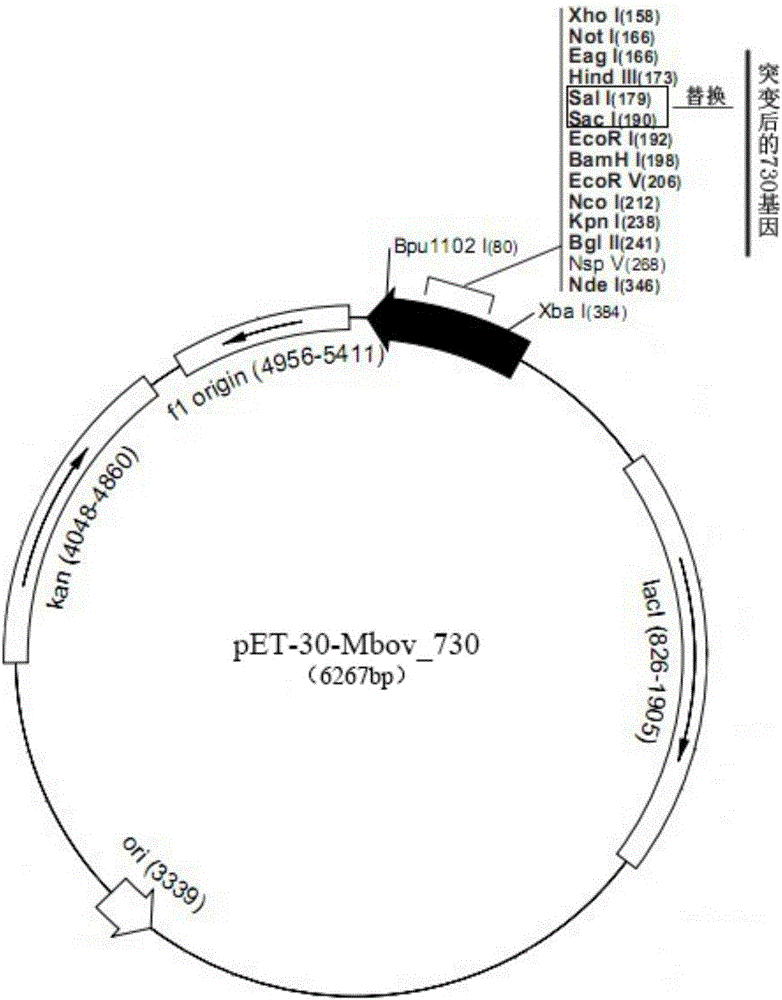
Application of mycoplasma bovis MbovP730 protein in natural infection and vaccine immunity identification
Owner:HUAZHONG AGRI UNIV
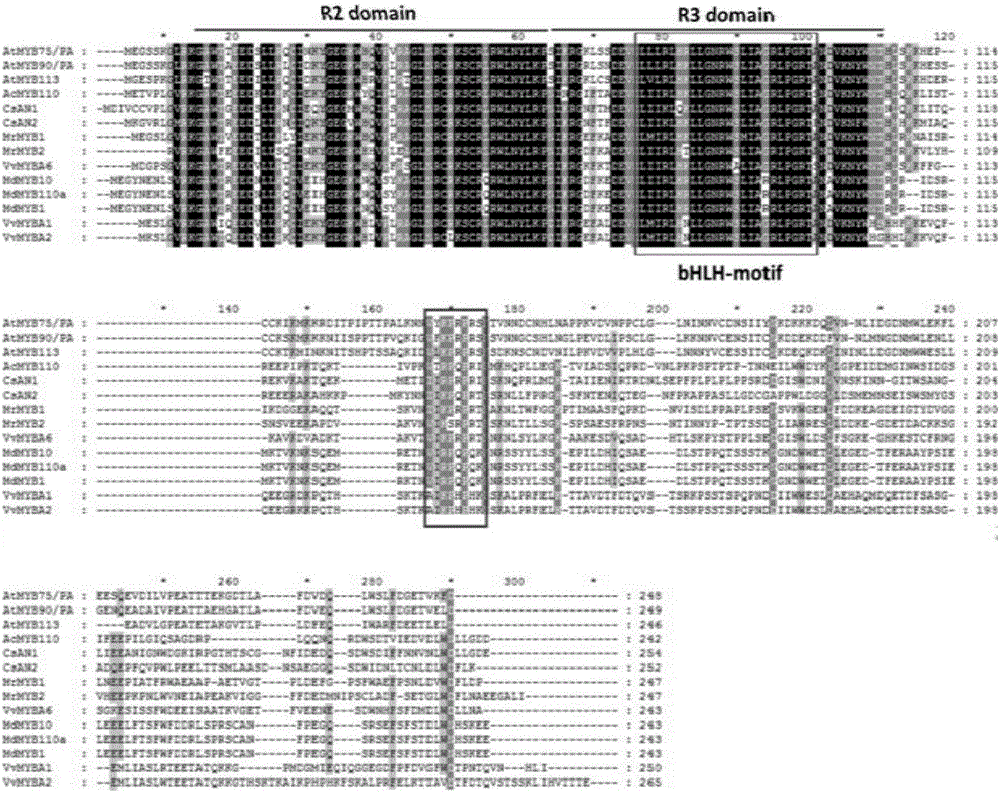
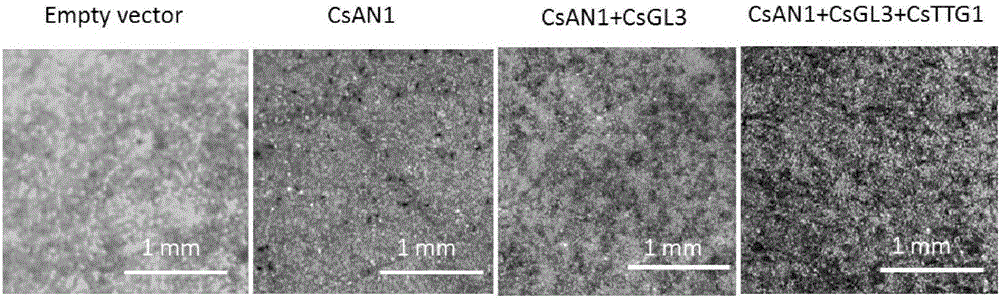
Tea tree MYB transcription factor CsAN1 and application thereof in regulation of anthocyanin metabolism
Owner:SOUTH CHINA AGRI UNIV
DNA bar code used for screening high-quality Tibet brown mushrooms, primer and application of DNA bar code used for screening high-quality Tibet brown mushrooms and primer
ActiveCN111235294AOvercome the shortcomings of not being accurate enough, time-consuming and labor-intensiveShort screening cycleMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyNucleotide
The invention discloses a DNA bar code used for screening high-quality Tibet brown mushrooms, a primer and application of the DNA bar code used for screening the high-quality Tibet brown mushrooms andthe primer. The DNA bar code used for screening the high-quality Tibet brown mushrooms contains one or more of 17 DNA fragments with nucleotide sequences shown in SEQ ID NO: 1-17 as shown in the description. The amplification primer of the DNA bar code used for screening the high-quality Tibet brown mushrooms contains one or more of 17 pairs of primers with upstream and downstream nucleotide sequences respectively shown in SEQ ID NO: 18-51 as shown in the description. Compared with a traditional breeding method and other existing DNA bar code technologies, the DNA bar code used for screeningthe high-quality Tibet brown mushrooms, the primer and application of the DNA bar code used for screening the high-quality Tibet brown mushrooms and the primer have the advantages of being time-saving, energy-saving, money-saving, accurate and efficient, and play a positive role in genetic breeding of the high-quality Tibet brown mushrooms, and an effective method is provided for identification and protection of germplasm resources at the same time.
Owner:LHASA PLATEAU BIOSES RES INST
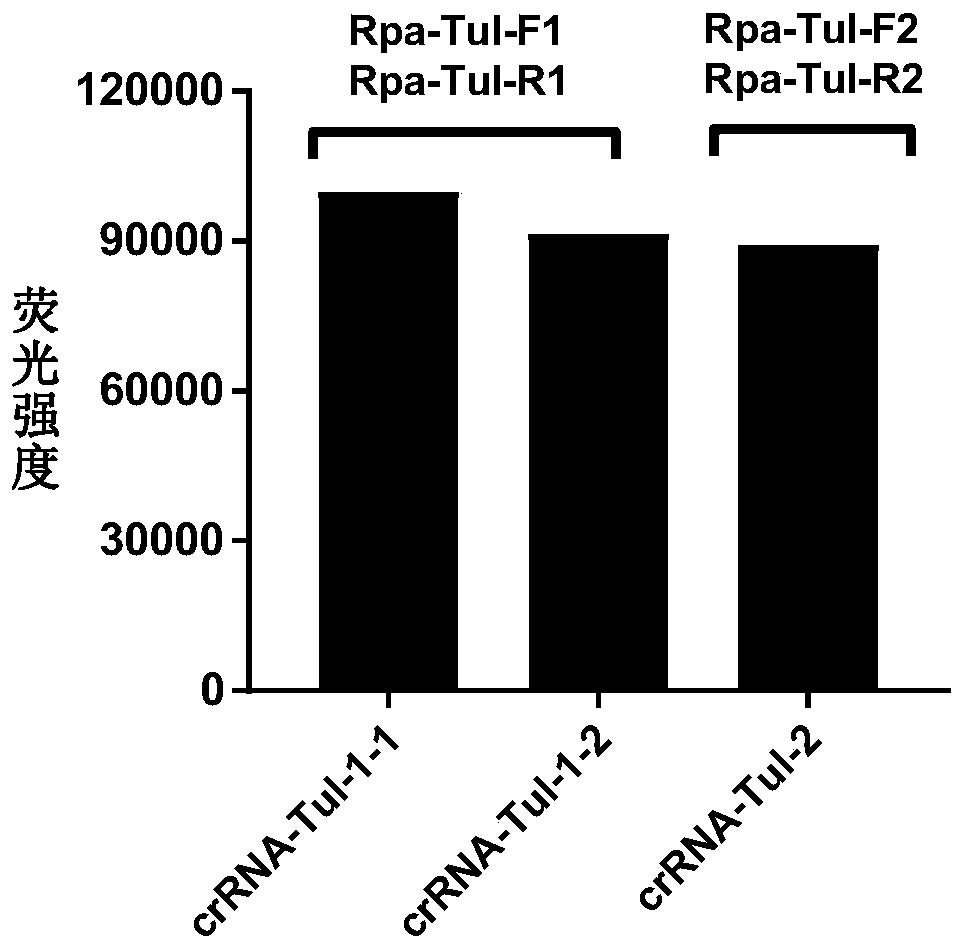
CRISPR-Cas12a detection primer group for francisella tularensis and application thereof
ActiveCN111394490AShort detection timeHigh amplification efficiencyMicrobiological testing/measurementMicroorganism based processesGeneticsNucleotide sequencing
Owner:ACADEMY OF MILITARY MEDICAL SCI
GTP cyclohydrolase I gene folE and application
Owner:KUNMING UNIV OF SCI & TECH
HCV (hepatitis C virus) core antigen and antibody thereof as well as hybridoma cell lines secreting antibody
ActiveCN102329378AGood antigenicityHigh purityVirus peptidesMicroorganism based processesAntigenNucleotide
Owner:SHANDONG UNIV QILU HOSPITAL
Screening and application of nucleic acid aptamer specifically targeting breast cancer cell line MCF-7
The invention discloses screening and application of a nucleic acid aptamer specifically targeting a breast cancer cell line MCF-7. The nucleotide sequence of the nucleic acid aptamer refers to a DNAfragment represented by the following sequence: a sequence 9-A: 5'-AGGAGCACGACTCTGACGTAGGATCGAGACGAGGTACGTAT-3'. The screening employs a cell-SELEX technology and comprises the following steps: firstsynthesizing an initial library; using the breast cancer cell line MCF-7 as target cells, and conducting incubation with the library; and conducting positive screening, elution, PCR amplification andsingle-stranded DNA preparation. For breast cancer cell detection, the nucleic acid aptamer has the advantages of simple and convenient operation, low cost, short cycle, and high accuracy; and targetmolecules combined with the nucleic acid aptamer of the invention are used for research, and tumor markers can be obtained. An effective tool and means are provided for early diagnosis of breast cancer, breast cancer typing and other applications, and the nucleic acid aptamer can be used for early diagnosis of breast cancer, preparation of breast cancer diagnostic drugs and kits.
Owner:GUOKE (JILIN) INTELLECTUAL PROPERTY OPERATION CO LTD
Polyphosphate kinase gene and applications thereof in sewage dephosphorization
The invention relates to a polyphosphate kinase gene and applications of the polyphosphate kinase gene in sewage dephosphorization, and particularly relates to three novel polyphosphate kinase genes,namely, ppk33, ppk44 and ppk73 which are respectively provided with nucleotide sequences as shown in SEQ NO 1, SEQ NO2 and SEQ NO.3. An engineering strain prepared by adopting the polyphosphate kinasegenes ppk33, ppk44 and ppk73 can be used for removing phosphorus element in sewage, wherein the strain M15 / pQE30a-ppk44T has the phosphorus removal amount being 7 times of that of a corresponding contrast strain, and the phosphorus removal efficiency achieves 70% after 10h.
Owner:HAINAN NORMAL UNIV
Probe composition for detecting lung cancer mutant genes based on NGS method and kit
ActiveCN110791500AStrong specificityHigh and uniform coverageMicrobiological testing/measurementDNA/RNA fragmentationNucleotideBlood plasma
The invention discloses a probe composition for detecting lung cancer gene mutation based on an NGS method and a kit thereof. The probe composition is selected from at least one of probes with nucleotide sequences as shown in SEQ ID NO.1-75, the kit is suitable for lung cancer gene mutation detection of FFPE, tissue and peripheral blood ctDNA based on the NGS method, and then the purposes of earlyscreening of lung cancer mutant genes, real-time monitoring of recurrence and the like are achieved. The uniquely designed UMI bimolecular tag can effectively reduce background noise, eradicate tracepollution, remove false positive and ensure the accuracy of a result, so that the sensitivity in ctDNA detection reaches 0.1%. A universal Short-Y joint is used in tissue detection, and the detectionsensitivity can reach 2%. The more possibilities are provided for accurate targeted therapy of patients. Tissue samples and plasma samples are similar in library building workflow, the simplicity ofthe workflow is guaranteed, time is saved, the efficiency is high, and the operation is easy.
Owner:KEAN BIOTECHNOLOGY (DALIAN) CO LTD
Callery pear ascorbate peroxidase gene and use thereof in resisting heavy metal stress
The invention relates to a callery pear ascorbate peroxidase gene and a use thereof in resisting heavy metal stress. The gene has a nucleotide sequence shown in the sequence SEQ ID No. 1. The total RNAs of the leaves of the callery pear are extracted after cadmium treatment and the ascorbate peroxidase gene Pc. APX is cloned by combination of bioinformatics and PCR so that the complete coding genesequence of 753 bp is obtained. The escherichia coli expression vector pET-22b(+)-Pc. APX is constructed and through the escherichia coli heterologous expression system, the functions of the cloned ascorbate peroxidase gene Pc. APX are identified and the recombinant escherichia coli with the cloned ascorbate peroxidase genes Pc. APX have strong tolerance to cadmium. At the same time, a binary plant expression vector pRI201-AN-GUS-Pc. APX is contructed, the vector is transferred into agrobacterium tumefaciens GV3101 cells by a freezing-thawing method, and the cells are transformed into Arabidopsis thaliana, and the obtained transgenic Arabidopsis thaliana has higher cadmium tolerance than wild-type Arabidopsis thaliana.
Owner:JIANGSU ACAD OF AGRI SCI
SNP (single nucleotide polymorphism) marker related to Erhualian sow litter traits and detection method and application thereof
ActiveCN104694651AImportant benefitsImportant social valueMicrobiological testing/measurementDNA/RNA fragmentationForward primerLitter
Owner:NANJING AGRICULTURAL UNIVERSITY
Method for producing l-amino acids
InactiveUS20090209012A1Efficient accumulationSatisfactory productivityBacteriaEnzymesHigh concentrationNucleotide
Owner:DAICEL CHEM IND LTD
Rice secretary type thioredoxin gene and application thereof
Owner:HEBEI NORMAL UNIV
One group of nucleic acid aptamers capable of specifically identifying Beijing genotypes tuberculosis bacterial strain antigen and application of nucleic acid aptamers
ActiveCN104450724AHigh affinityStrong specificityMaterial analysisDNA/RNA fragmentationAptamerAntigen
Owner:武汉顺可达生物科技有限公司
Overexpression of Plant Mirnas for Parasite Control
InactiveUS20140007295A1Reduce formationClimate change adaptationOther foreign material introduction processesHeterologousParasite Control
Owner:SYNGENTA PARTICIPATIONS AG
Molecular marker for identifying rice grain length characters, identification method and application
InactiveCN107245530AEasy to operateReduce consumptionMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceNucleotide
Owner:LIAONING PROVINCIAL SALINE ALKALI LAND UTILIZATION & RES INST
Diamondback moth cecropin 3, preparation method and application thereof
Owner:SOUTH CHINA AGRI UNIV
Absolute fluorescence quantitative PCR (Polymerase Chain Reaction) primer and kit for detecting atypical porcine pestivirus
InactiveCN107513583AEasy to operateStrong specificityMicrobiological testing/measurementMicroorganism based processesNucleotideFluorescence
Owner:SOUTH CHINA AGRI UNIV
Nucleic acid aptamer capable of detecting human colon cancer and application thereof in preparing detection preparations
ActiveCN109628455AAccurate diagnosisRapid positioningMaterial analysisDNA/RNA fragmentationChemical synthesisAptamer
The invention discloses a nucleic acid aptamer capable of achieving targeted detection of colon cancer cells and application thereof. The nucleotide sequence of the nucleic acid aptamer is 5'-ACGCTCGGATGCCACTACACGGTTGGGGTCGGGCATGCGTCCGGAGAAGGGCAAACGAGAGGTCACCAGCACGTCCATGAG-3'. The nucleic acid aptamer is good in stability, target molecules can be specifically identified, the immunogenicity in thebody is small, and the target molecules can be easily removed. The aptamer is small in molecular weight and low in preparation cost, can be obtained through chemical synthesis in vitro, and is easy tostore and transport. By adopting the nucleic acid aptamer, various colon cancer cells can be detected, the operation is simple and quick, and early diagnosis, targeted treatment, prognosis and the like of colon cancer are facilitated.
Owner:HUNAN UNIV
Kit for detecting human herpes virus infection and detection method thereof
PendingCN111500788AEasy to detectQuick checkMicrobiological testing/measurementDNA/RNA fragmentationInfected patientNucleotide
Owner:领航医学科技(深圳)有限公司
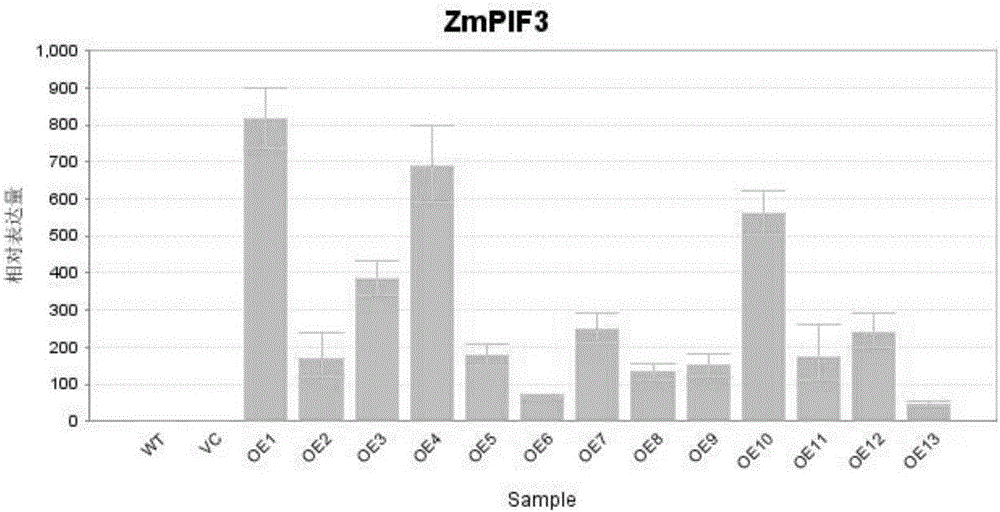
Molecular marker of maize transcription factor ZmPIF3 transgenic rice and application of molecular marker
InactiveCN105695459AImprove success efficiencyGood repeatabilityMicrobiological testing/measurementDNA/RNA fragmentationGenetically modified riceAgricultural science
Owner:YANGZHOU UNIV
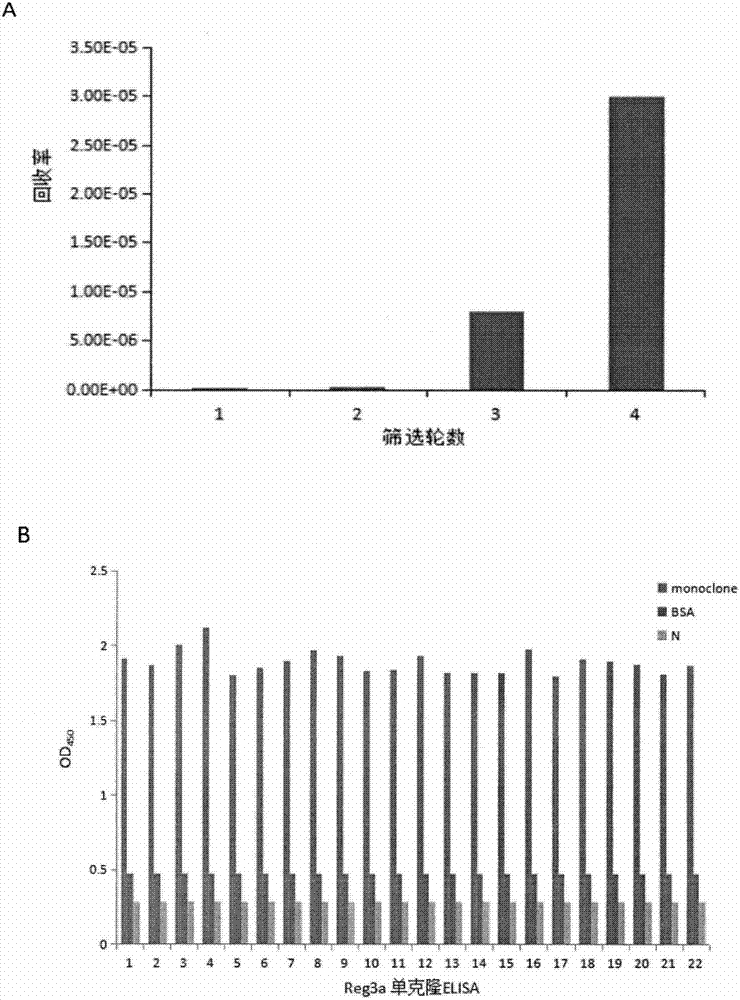
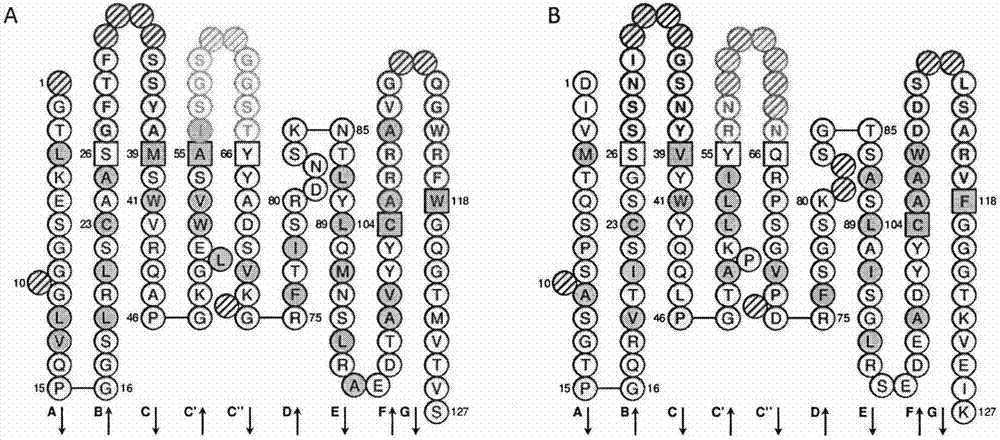
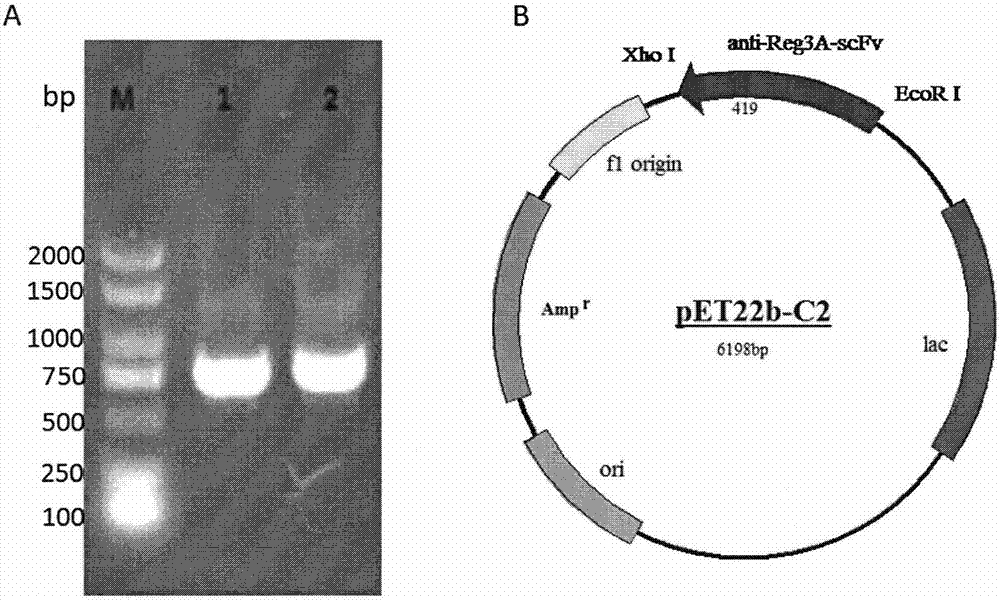
Single-chain antibody for targeting Reg3A
InactiveCN107365384APromote application developmentGreat advantageBacteriaMicroorganism based processesSingle-Chain AntibodiesNucleotide
Owner:CHINA PHARM UNIV
Method for detecting marine products of same origin
The invention provides a method for detecting marine products of the same origin. The method comprises the following steps: designing a specific primer by utilizing a CO1 sequence of cytochrome oxidase genes in mitochondrial genes of a group of marine products of the origin, performing PCR amplification on the different extracted marine products, and generating nucleotide sequences of different PCR fragments; comparing different gene sequences with the CO1 gene sequence of the marine products of the origin, performing biological analysis, thereby determining the authenticity of the products. A rapid, accurate and sensitive authentication method is established for protecting the geographical indication products. According to collection and amplification comparison of a group of conserved genes in aquatic products, molecular bar codes are marked on the geographical indication aquatic products, so that the origin is identified, the local fishery resources are protected, and the competitiveness and popularity of local fishery are improved.
Owner:OCEAN RES CENT OF ZHOUSHAN ZHEJIANG UNIV
Scylla paramamosain C-type lysozyme gene and application thereof
ActiveCN109385436AHas inhibitory effectAntibacterial agentsPeptide/protein ingredientsScylla paramamosainLysobacter enzymogenes
The invention discloses a Scylla paramamosain C-type lysozyme gene, the nucleotide sequence of the gene is shown in sequence table SEQ ID No: 1. The amino acid sequence of the coding protein of the Scylla paramamosain C-type lysozyme gene is shown in SEQ ID No: 2. The invention also discloses application of the coding protein of the Scylla paramamosain C-type lysozyme gene in preparation of medicaments for inhibiting bacteria.
Owner:SOUTH CHINA SEA FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
Neural cells expressing adenovirus E4ORF1, and methods of making and using the same
The present invention discloses engineered neural cells, neural stem cells, or neural progenitor cells that contain a nucleotide sequence that encodes an adenovirus E40RF1 polypeptide and / or that contain an adenovirus E40RF1 polypeptide. The present invention also provides methods of making and using the engineered cells and compositions comprising the engineered cells.
Owner:ANGIOCRINE BIOSCI
Primer group used for detecting canine parvovirus and application of primer group used for detecting canine parvovirus
PendingCN111394512APracticalStrong specificityMicrobiological testing/measurementMicroorganism based processesNucleotideCanine parvovirus
The invention discloses a primer group used for detecting a canine parvovirus. The primer group comprises an outer primer pair, an inner primer pair and an annular primer pair, wherein a nucleotide sequence of one primer in the outer primer pair is shown in SEQ ID NO:1 as shown in the description, and a nucleotide sequence of the other primer in the outer primer pair is shown in SEQ ID NO:2 as shown in the description; a nucleotide sequence of one primer in the inner primer pair is shown in SEQ ID NO:3 as shown in the description, and a nucleotide sequence of the other primer in the inner primer pair is shown in SEQ ID NO:4 as shown in the description; and a nucleotide sequence of one primer in the annular primer pair is shown in SEQ ID NO:5 as shown in the description, and a nucleotide sequence of the other primer in the annular primer pair is shown in SEQ ID NO:6 as shown in the description. According to the primer group used for detecting the canine parvovirus, according to the sequence characteristics of a VP2 gene in the genome of the canine parvovirus, the specific real-time fluorescence loop-mediated isothermal amplification primer group is designed, the practicability is high, a result can be directly judged by means of real-time fluorescence signals and a melting curve, the specificity is high, and the sensitivity is high.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI
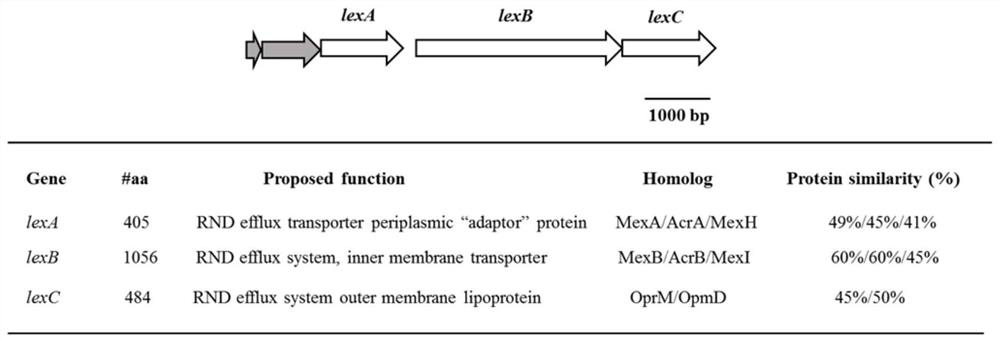
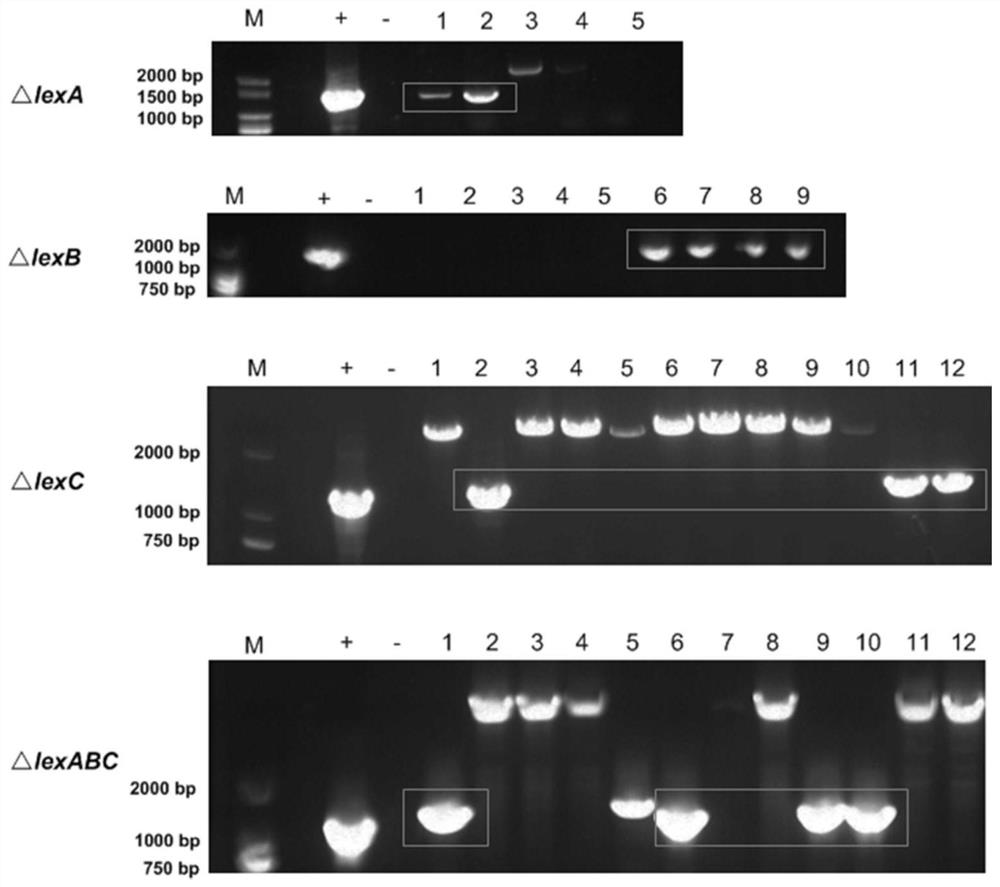
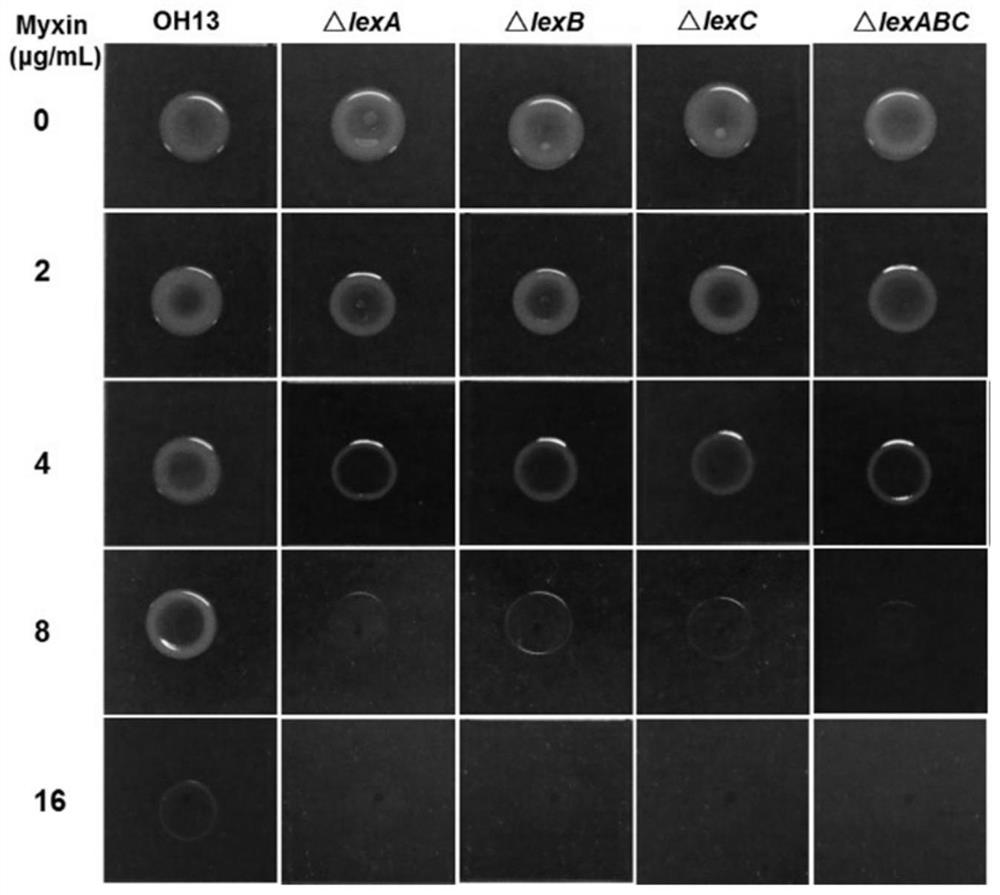
Efflux pump LexABC participating in self resistance of phenazine substance myxin as well as coding gene and application of efflux pump LexABC
PendingCN113186201AIncrease resistanceFree from toxicityBacteriaMicroorganism based processesBiotechnologyLysobacter antibioticus
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Panax notoginseng reverse osmosis protein gene PnOLP1 and application
ActiveCN108707611AIncrease resistanceReduce usagePlant peptidesFermentationGenomicsNigrospora oryzae
Owner:KUNMING UNIV OF SCI & TECH
Primer, kit and identification method for identifying bacillus amyloliquefaciens in yeast for making hard liquor
PendingCN114540519ARapid identificationRapid responseMicrobiological testing/measurementMicroorganism based processesBiotechnologyYeast
Owner:SICHUAN UNIV
Who we serve
- R&D Engineer
- R&D Manager
- IP Professional
Why Eureka
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Social media
Try Eureka
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap