Primer, kit and identification method for identifying bacillus amyloliquefaciens in yeast for making hard liquor
A technology for amylolytic bacillus and bacillus, which is applied in the field of liquor detection, can solve the problems of large error, time-consuming, and difficulty in distinguishing the forms of Bacillus close relatives, etc., and achieve the effect of fast response, easy operation, and accurate qualitative identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1: Design of ARMS primers
[0043](1) Amplification of Bacillus amyloliquefaciens using primers 2-F and 2-R
[0044] Primer Nucleotide Sequences:
[0045] 2-F: TCGGAAGTGCGGCTGGATC (SEQ ID NO: 3)
[0046] 2-R: AAGGCATCCACCGTGCGCCC (SEQ ID NO: 4)
[0047] Amplified Bacillus amyloliquefaciens sequence:
[0048] CTAGTAAGTTGTCTGCCAGATTACGGATATAAGACCTTGGGTCTTATAAACAGAACGTTCCCTGTCTTGTTTAGTTTTGAAGGATCATTCGATTCTTCGAGATAATGTTCTTTGAAAACTAGATAACAGAAGTAATTCACATTCAATTGTAATGCAAGATATCACGTAGTGATTCTTTTTAACGGTTAAGTTAGAAAGGGCGCACGGTGGATGCCTTA. (SEQ ID NO: 5)
[0049] Copy this sequence into the National Center for Biotechnology Information (NCBI) official website to get a longer sequence:
[0050] ACAGATGATTGGGGTGAAGTCGTAACAAGGTAGCCGTATCGGAAGGTGCGGCTGGATCACCTCCTTTCTAAGGATTTTAACGGAATATAAGACCTTGGGTCTTATAAACAGAACGTTCCCTGTCTTGTTTAGTTTTGAAGGATCATTCGATTCTTCGAGATGTTGTTCTTTGAAAACTAGATAACAGAAGTAATTCACATTCAATTGTAATGCAAGATATCACGTAGTGATTCTTTTTAACGGTTAAGTTAGAAAGGGCGCACGGTGGATGCCTTGGCACTAGGAGC
Embodiment 2
[0068] Example 2: Extraction of strain genomic DNA
[0069] (1) Liquid culture of strains: Inoculate porcelain beads of 8 strains from porcelain bead storage tubes and put them into liquid medium under aseptic conditions.
[0070] Liquid medium: 5 g of yeast extract, 10 g of peptone, 10 g of sodium chloride, pH 7.0-7.2, 1 L of distilled water, sterilized at 121 °C for 30 min.
[0071] (2) Aspirate 1 mL of overnight bacterial solution, centrifuge to obtain bacterial cells, use Ezup column bacterial gene DNA extraction kit to extract DNA, and store at -20°C for future use.
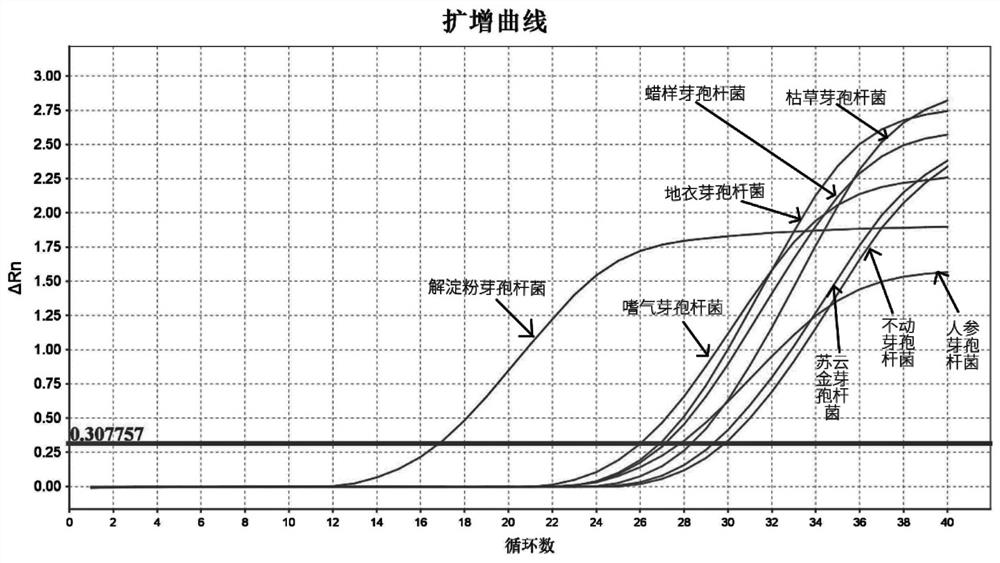
[0072] The experimental strains include: Bacillus amyloliquefaciens, Bacillus subtilis, Bacillus licheniformis, Bacillus cereus, Bacillus thuringiensis, Bacillus aerophilus, Bacillus acuminata, Bacillus ginseng.
Embodiment 3
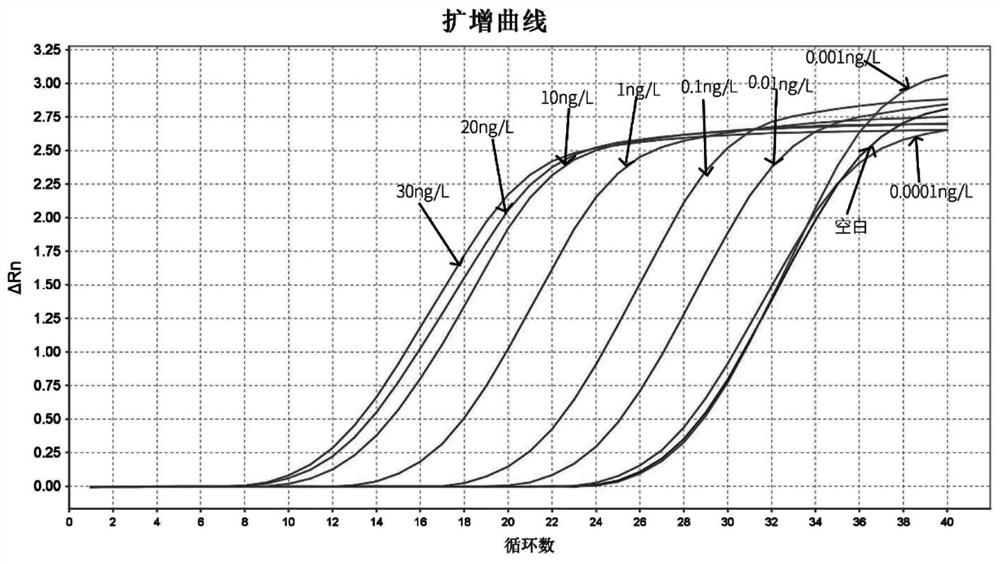
[0073] Example 3: Verify the validity of 8 pairs of ARMS primers
[0074] (1) Establishment of real-time fluorescent quantitative PCR method:
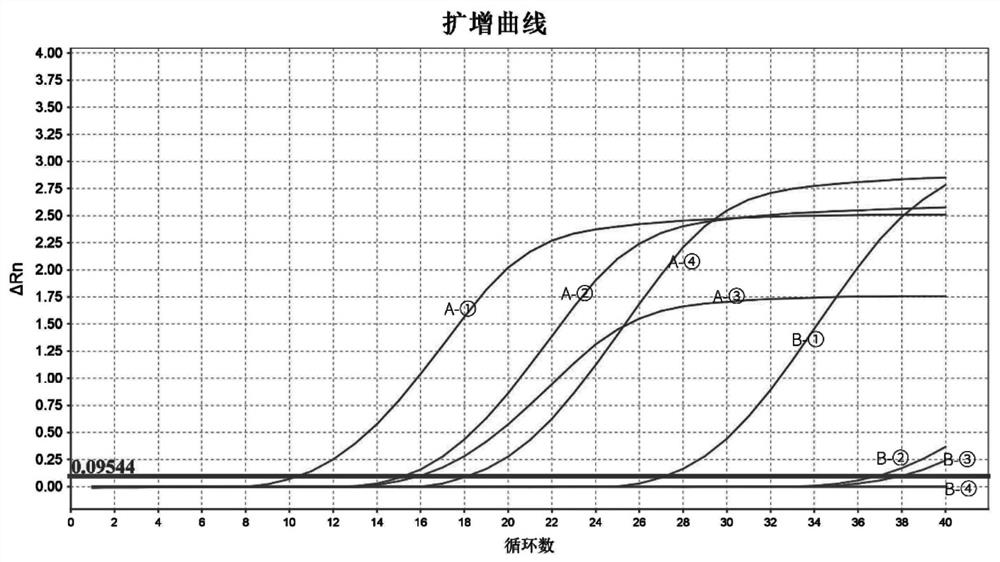
[0075] Total volume 20μL, including 10μL qPCR Mix, 0.8μL upstream and downstream primers (2.5 μmol / L), 1μL DNA template, ddH 2 O7.4uL. The DNA templates were the extracted DNA of Bacillus amyloliquefaciens and Bacillus subtilis.
[0076] It should be noted that, because ① the common strains producing tetramethylpyrazine in Daqu are mainly Bacillus licheniformis, Bacillus amyloliquefaciens and Bacillus subtilis, etc.; ② the applicant compared the bases from the NCBI gene bank in the early stage. It is found that only the base sequences of Bacillus subtilis and Bacillus amyloliquefaciens are very similar, which is the most difficult to distinguish; the base sequences of other strains such as Bacillus licheniformis and Bacillus amyloliquefaciens are relatively different and easier to distinguish. Therefore, the first step to verify the
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap