Method for detecting plasmodium ovale infection by using PCR technology
A Plasmodium ovale detection primer technology, applied in the field of molecular biology, can solve the problems of high misdiagnosis rate, difficult identification between species, difficulty in detection of Plasmodium ovale infection, etc., and achieve the effect of strong specificity and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Example Embodiment
[0017] The design of specific primers: Example 1
[0018] Experimental material
[0019] Synthesis and nucleotide sequencing primers by Suzhou intellectualism Biotechnology Co., Ltd. completed.
[0020] 2. primer design
[0021] The GenBank database sequence encoding ovale (as shown in SEQ ID NO.1) and Plasmodium vivax and Plasmodium falciparum (P. falciparum: Gene ID: 3885775, Plasmodium vivax: GeneID: 5475443) in homologous sequence comparison, according to the difference sites designed sequence comparison of the PCR obtained ovale detection primer, designed to give the primers were synthesized by Suzhou intellectualism biotechnology company, specific detection of PCR primer sequences were as follows,
[0022] Primers were as follows:
[0023] SEQ ID NO2: ATGGTAGGGGCTAAAGGAGTG, SEQ ID NO.2;
[0024] SEQ ID NO3: CAATATGGTGAA AAAAACTGAGGAC, SEQ ID NO.3.
Example Embodiment
[0025] Example 2: PCR amplification of the target gene from a test sample
[0026] Experimental material
[0027] Genomic DNA extraction kit purchased from Tiangen Technology (Beijing) Company, Agarose TransGen powder available from the company, PCR instrument available from (Eppendorf).
[0028] 2. Extraction of genomic DNA to be detected in the serum
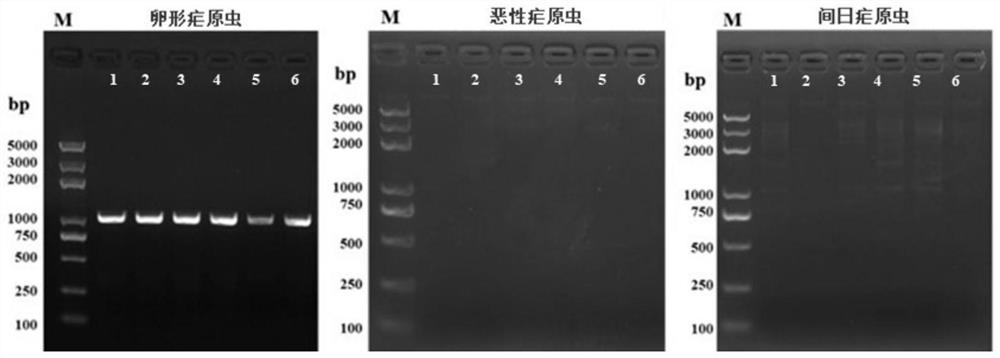
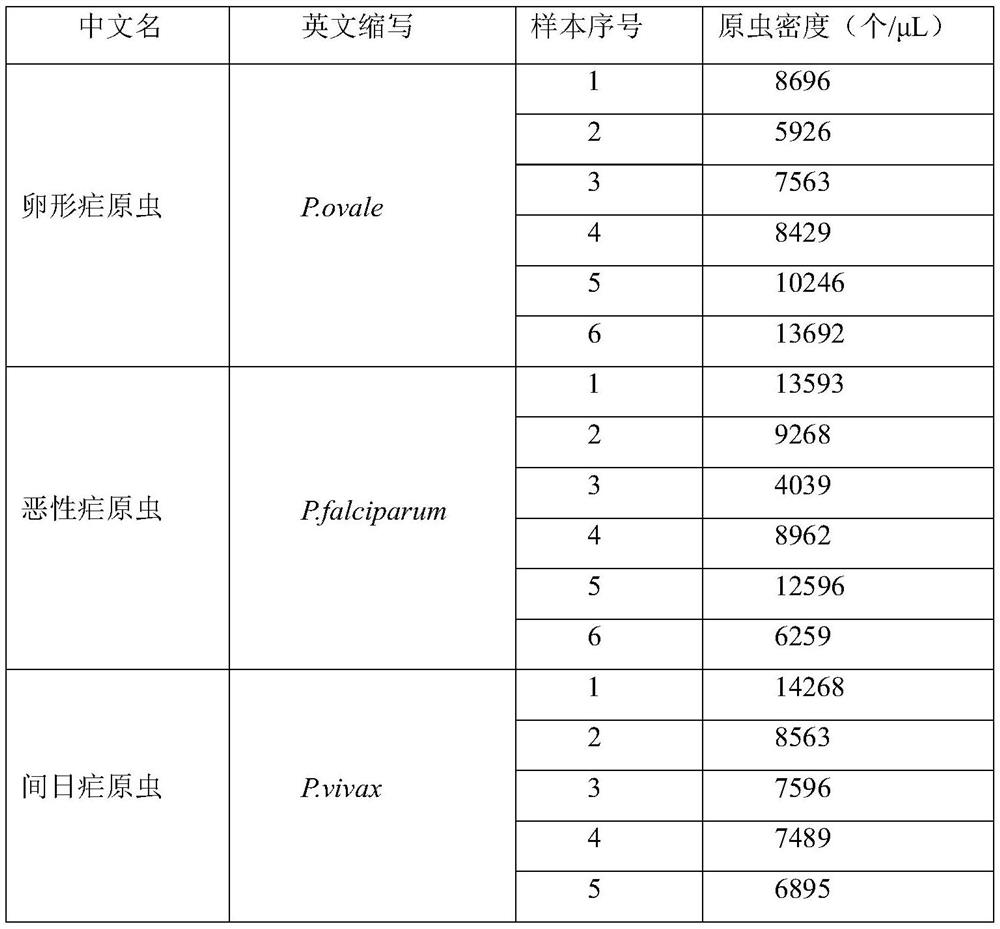
[0029] They were taken Plasmodium ovale, Plasmodium falciparum and Plasmodium vivax six patients in each of the DNA samples extracted using Tiangen Technology (Beijing)'s blood / cell / tissue genomic DNA extraction kit, according to the method description parasitemia level of genomic DNA genomic DNA, extracted, and the extracted DNA samples provided at -20 ℃ refrigerator for use, the samples were as shown in table 1.
[0030] 3.PCR amplified specific bands
[0031] Use Suzhou intellectualism biotechnology companies synthesized primers specific to the genomic DNA samples extracted from amplified verified, the PCR instrument using
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap