Method for analyzing and mining functional genes by combining genome three-dimensional structure difference identification and transcriptome gene expression level difference
A technology of three-dimensional structure and identification method, which is applied in the field of identification of three-dimensional structure differences of genomes, and can solve problems such as the small number of differential loops.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1 Identification method of genome three-dimensional structure difference
[0063] In this embodiment, the Hi-C data of chordoma and normal samples (two biological replicates for each sample, and the two samples are named H01 and H02 respectively) are used as the analysis object to provide a method for identifying differences in the three-dimensional genome structure. details as follows:
[0064] 1. Acquisition of effective Hi-C data
[0065] Using bwa and HiC-Pro to compare and quality control the Hi-C data, only keep the valid Hi-C data, and finally obtain the interaction matrix with bin sizes of 100000, 20000 and 10000bp.
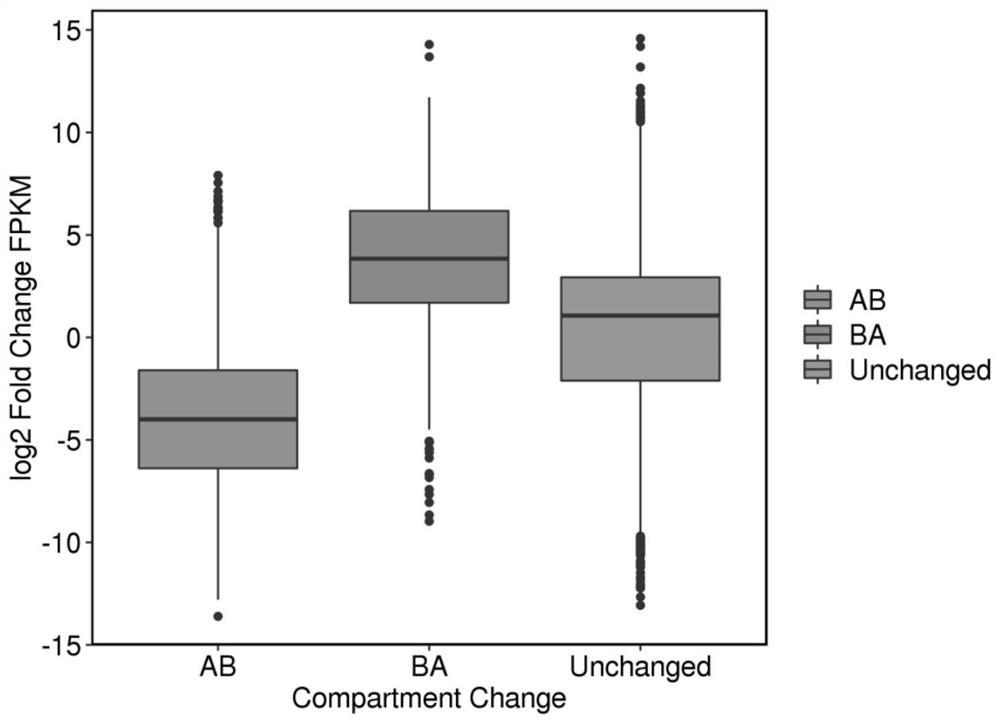
[0066] 2. Identification of difference AB Compartment
[0067] (1) Use HiTC and bin=100000 to obtain the identification result of AB Compartment for each sample;
[0068] (2) The identification of A and BCompartment is carried out separately for the biological repeats of each sample, and the consistent identification results among the biolo
Embodiment 2
[0084] Example 2 A functional gene mining method that combines the identification of genome three-dimensional structure differences and the analysis of transcriptome gene expression level differences
[0085] This example takes the identification results of genome three-dimensional structure differences in Example 1 and transcriptomics gene expression differential data of normal and chordoma samples as analysis objects, and provides a method that combines identification of genome three-dimensional structure differences and transcriptome gene expression level differential analysis The functional gene mining method is as follows:
[0086] (1) Use bedtools to correlate the differentially expressed genes with the AB and BA types in the differential AB Compartment to get the intersection, and obtain the differentially expressed gene information corresponding to the differential AB Compartment;
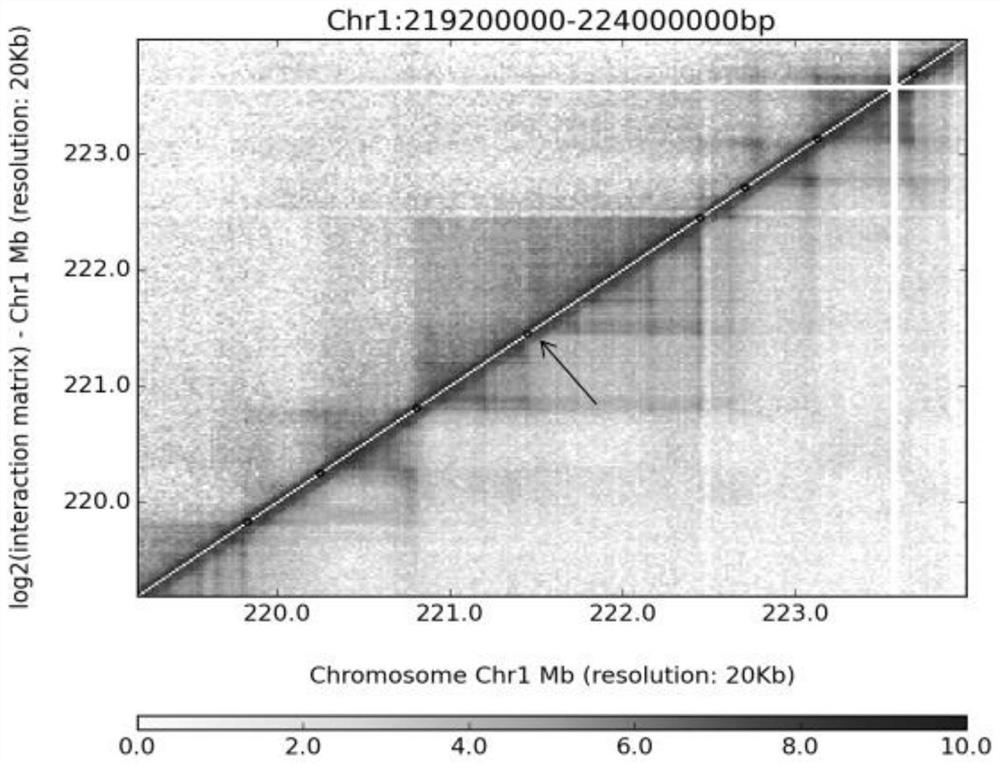
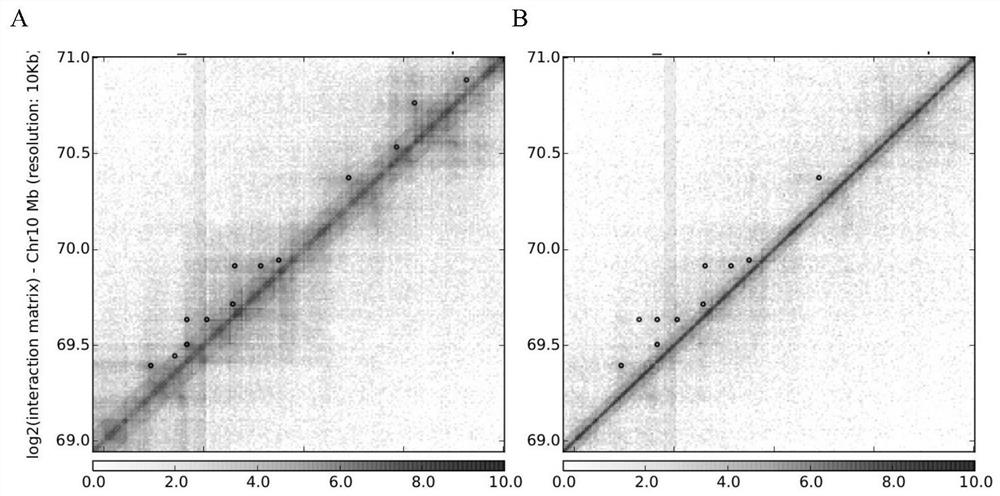
[0087] (2) Extend the boundary of the differential TAD by 40,000 bp to the inside and outs
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap