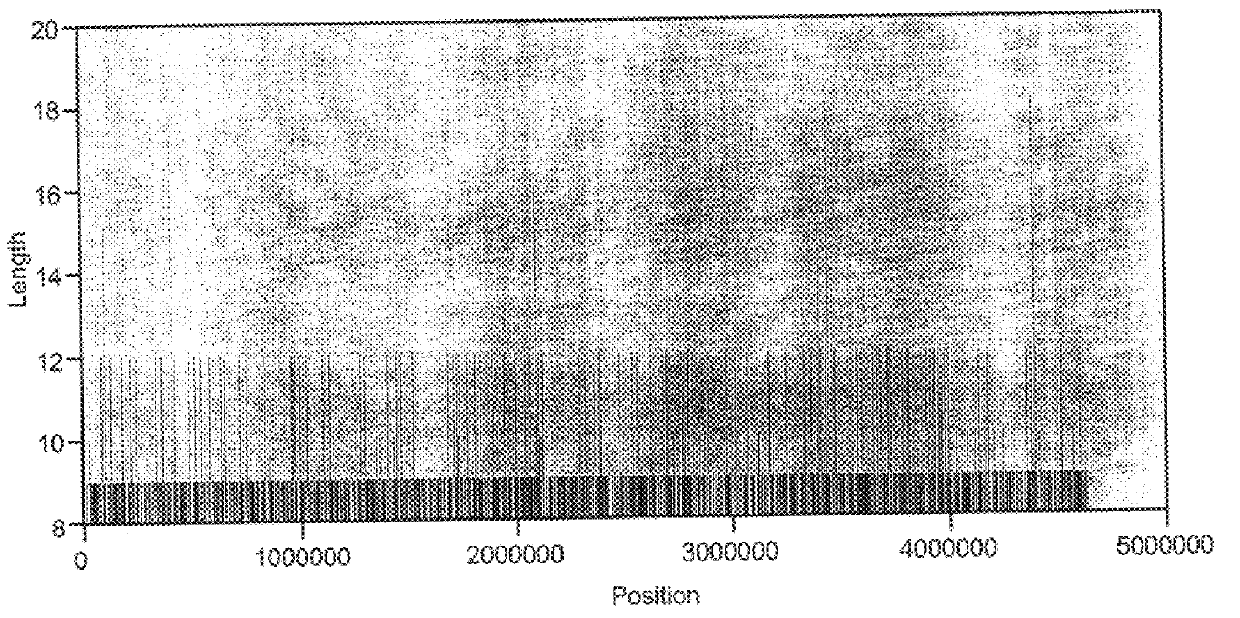
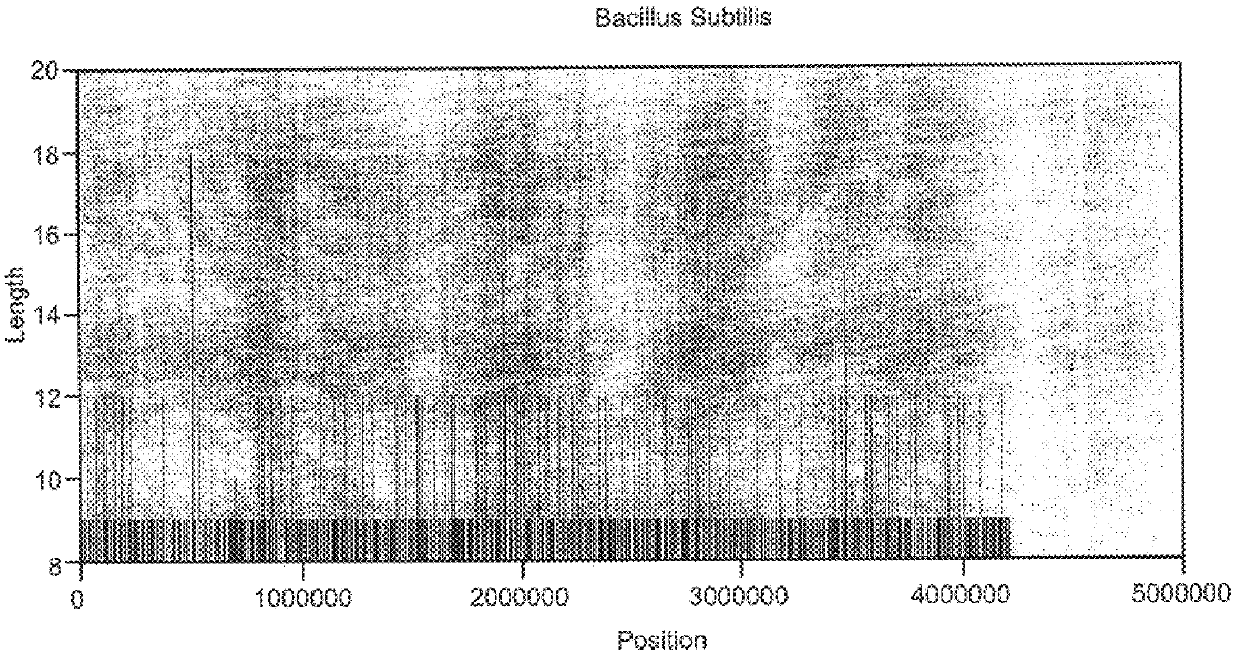
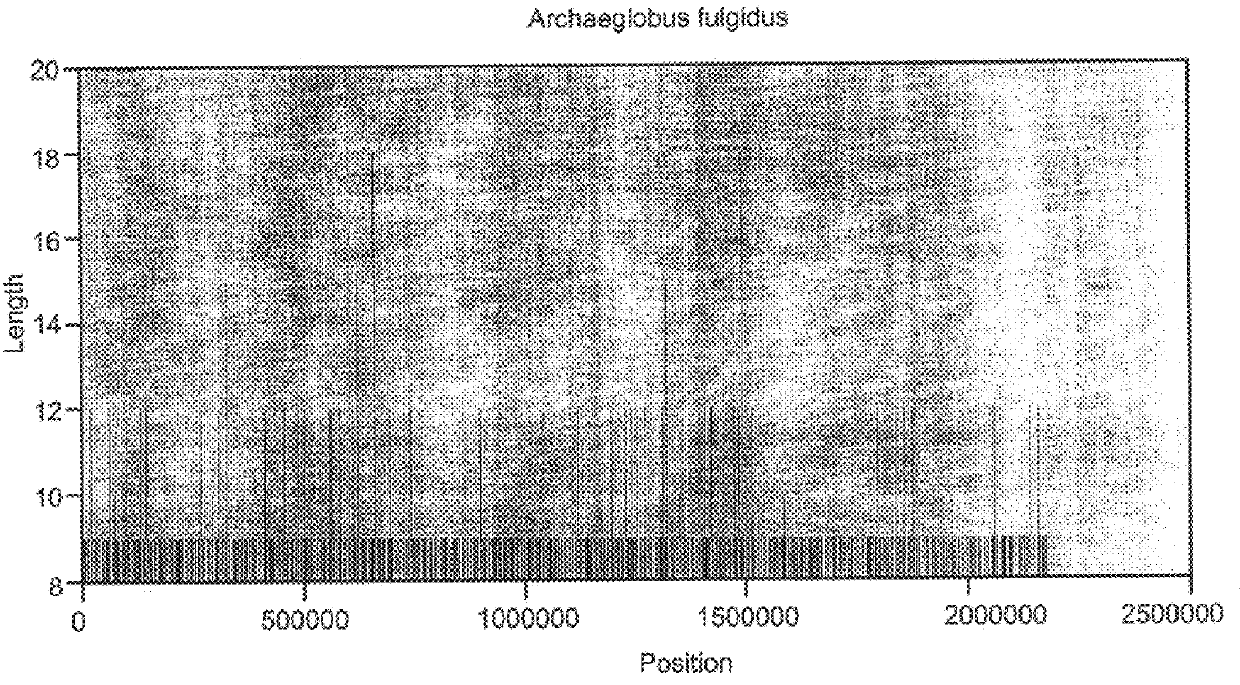
Abundant, well distributed and hyperpolymorphic simple sequence repeats in prokaryote genomes and use of same for prokaryote classification and typing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Example
Reference is now made to the following examples, which together with the above descriptions, illustrate the invention in a non limiting fashion.
Generally, the nomenclature used herein and the laboratory procedures in recombinant DNA technology described below are those well known and commonly employed in the art. Standard techniques are used for cloning, DNA and RNA isolation, amplification and purification. Generally enzymatic reactions involving DNA ligase, DNA polymerase, restriction endonucleases and the like are performed according to the manufacturers' specifications. These techniques and various other techniques are generally performed according to Sambrook et al., Molecular Cloning--A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. (1989). The manual is hereinafter referred to as "Sambrook". Other general references are provided throughout this document. The procedures therein are believed to be well known in the art and are provided for the convenien
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap