PCV2-PRV (porcine circovirus type 2-porcine pseudorabies virus) two-temperature type double PCR (polymerase chain reaction) detection primers and diagnosis method thereof
A diagnostic method, PCV2-F2 technology, applied in the biological field, can solve the problems of laborious, time-consuming, non-specific reaction and low sensitivity, and achieve the effect of low cost, simple operation and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] PCV2-PRV two-temperature double PCR detection primers, including PCV2 virus primer pair and PRV virus primer pair;
[0031] Wherein, the PCV2 virus primer pair includes upstream primer PCV2-F1 and downstream primer PCV2-F2; the PRV virus primer pair includes upstream primer PRV-F3 and downstream primer PRV-F4;
[0032] The sequence of the upstream primer PCV2-F1 is: 5'-AGTCTCATCCACAGCTGATTC-3';
[0033] The sequence of the downstream primer PCV2-F2 is: 5'-CATCTTGGCCAGATCCTCCG-3';
[0034] The sequence of the upstream primer PRV-F3 is: 5'-TCCTGTACGCCCTATTCA-3';
[0035] The sequence of the downstream primer PRV-F4 is: 5'-CCGACTCTCGTGATGTCATCC-3'.
Embodiment 2
[0037] The PCV2-PRV two-temperature double PCR diagnostic method comprises the following steps:
[0038] 1) Synthesize PCV2 virus primers to PCV2-F1 / PCV2-F2 and PRV virus primers to PRV-F3 / PRV-F4;
[0039] 2) Extracting viral genomic DNA from PCV2 and PRV disease materials;
[0040] 3) PCV2 DNA and PRV DNA were amplified and detected by two-temperature PCR.
[0041] Further, the method for extracting the viral genomic DNA in step 2) is as follows: take 0.1 g of PCV2 and PRV positive disease materials respectively, place them in an Eppendorf tube, add 1 ml of TE buffer solution, and twist it into a homogenate with a twister, - Freeze and thaw three times at 70°C, centrifuge to get 0.2ml of the supernatant, then use a viral DNA extraction kit to extract viral DNA, and finally elute in 50ul TE buffer and store at -20°C to obtain PCV2DNA and PRV DNA.
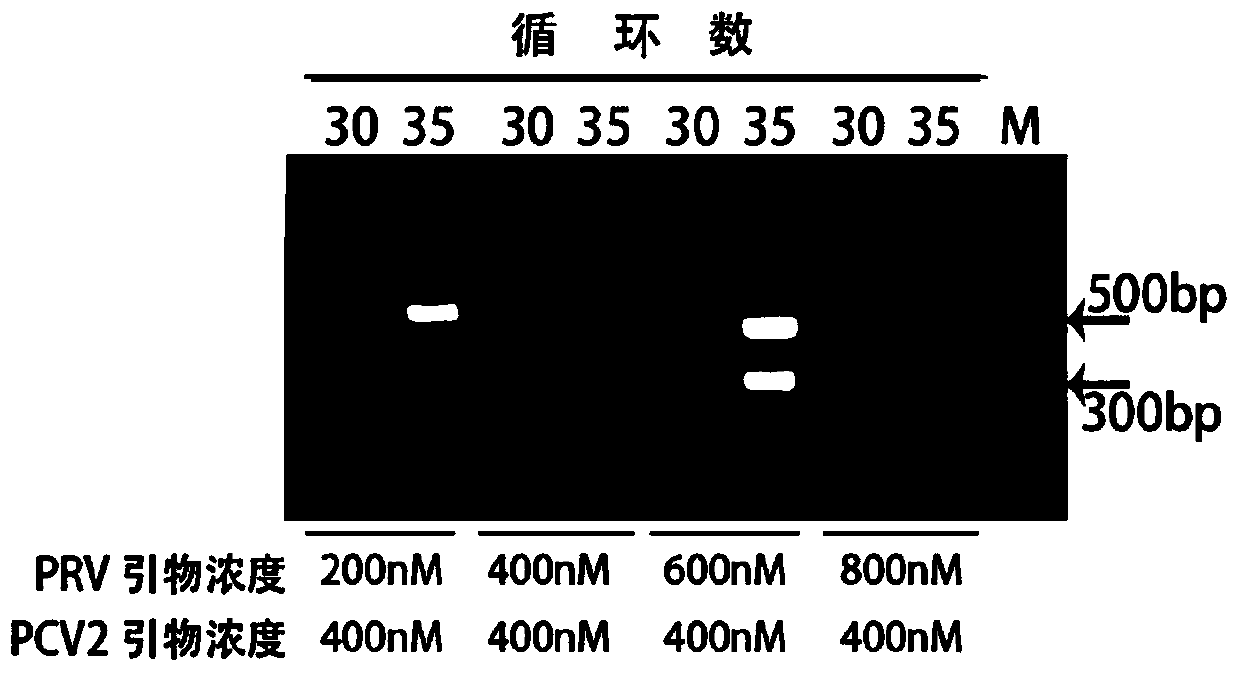
[0042] Further, the method of the two-temperature type PCR amplification in step 3) is:
[0043] Use PCV2 and PRV DNA as templ
Embodiment 3
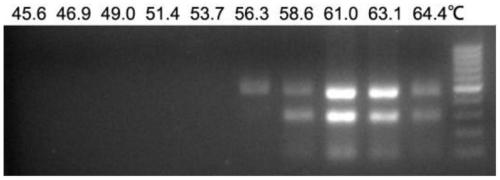
[0053] The method for determining the optimal annealing temperature for two-temperature PCR is: mix the 50 μl reaction system in Example 2 and put it in a PCR amplification instrument. The amplification conditions are: 95°C for 5 minutes; ℃, 46.9°C, 49.0°C, 51.4°C, 53.7°C, 56.3°C, 58.6°C, 61.0°C, 63.1°C, 64.4°C for 45s, 35 cycles; the final extension at 72°C for 10 min, after agarose gel electrophoresis, as figure 1 As shown, the results show that the optimal annealing temperature of PRV is 61°C, at which point the two specific bands can be clearly distinguished, and the product amplification efficiency is the highest.
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap