MNP marker site, primer composition and kit for cassava variety identification and application thereof
A technique of primer composition and marker site, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/testing, etc., can solve the problems of low polymorphism, poor flexibility, difficult adjustment and change, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1, be used for the screening of the MNP marker site of cassava variety identification and the design of multiple PCR amplification primers
[0038] S1. Screening of MNP marker loci for identification of cassava varieties
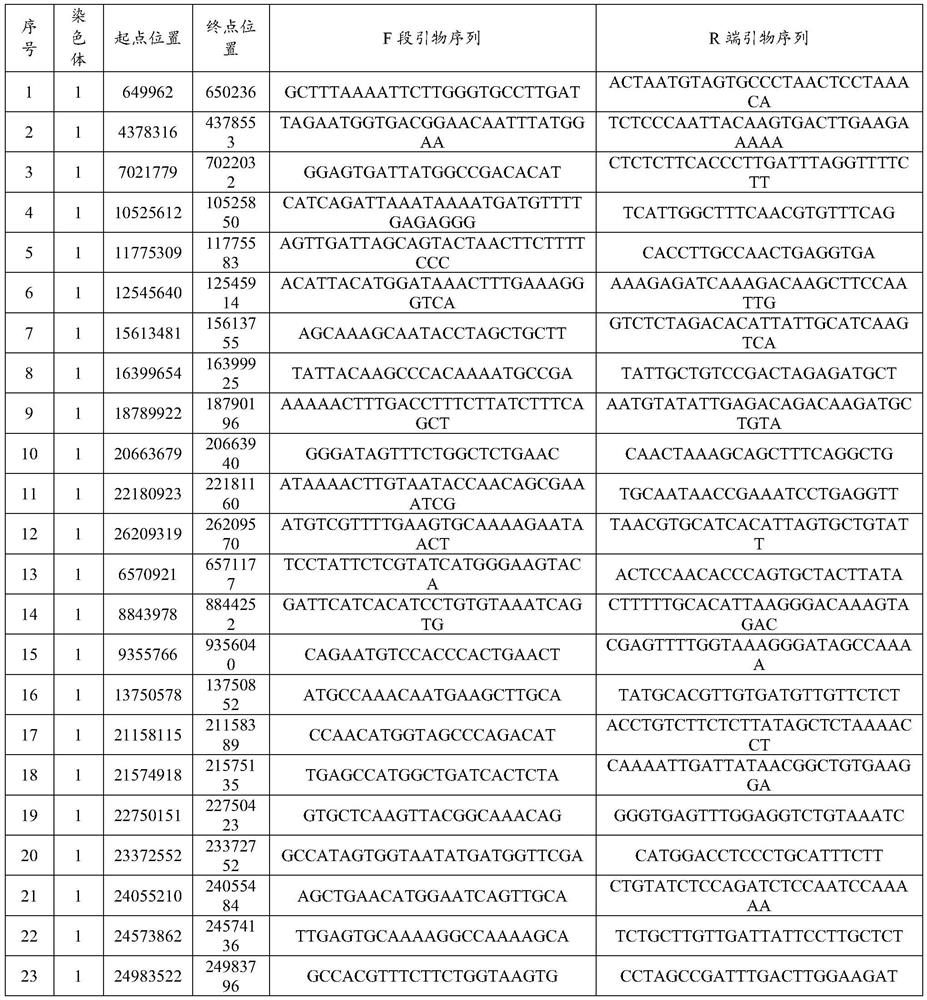
[0039] Taking the publicly released cassava genome sequence Mesculenta_305_v6 as the reference genome, combined with the genome sequencing data of 102 cassava varieties and resources collected by our research group, firstly use Samtools (Version 1.2) and BCFtools (Version: 1.2) for sequence analysis to obtain the cassava genome The SNP loci on the above, and compared with the NT library of NCBI, the screening of MNP markers was carried out according to the following principles: (1) The marker sequence is only unique to cassava and does not appear in other species; (2) The sequence has a single copy in the genome (3) there are at least three or more discontinuous SNP differences in the marker sequence; (4) the length of the marker sequence is l
Embodiment 2
[0062] Example 2, Evaluation of MNP markers for cassava variety identification, primer compositions and kits thereof
[0063] After 623 pairs of primers were synthesized, an equal amount of 5ul per primer was mixed to form a 1:1 primer mix of F and R-terminal primers. The 30 cassava varieties and resources collected by the unit were used to evaluate the developed MNP markers, primers and kits to test the detection rate, accuracy and discrimination of MNP marker loci.
[0064] (1) MNP marker detection rate
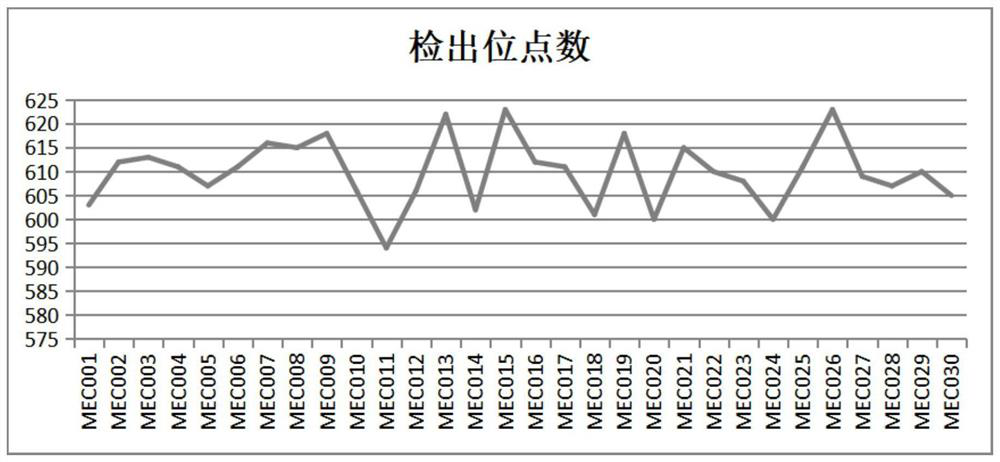
[0065] Carry out multiple PCR amplification and the construction of sequencing library according to the kit of the present invention, carry out multiple amplification, second-generation high-throughput sequencing and data analysis to these 30 cassava DNA samples, and detect 30*623= in one experiment simultaneously With 18,690 marker sites, the average sequencing coverage of each sample is more than 800 times, showing the high efficiency of MNP marker detection.
[0066] The
Embodiment 3
[0077] Embodiment 3, authenticity identification of cassava varieties and determination of substantive derived varieties
[0078] The current national standard of "Plant Variety Identification MNP Marking Method" uses the genetic similarity coefficient as the basis for variety identification, and the calculation of the genetic similarity coefficient is based on the number of marker loci with differences and the same genotype between varieties. , so the accuracy of breed identification ultimately depends on the accuracy of genotyping of breeds.
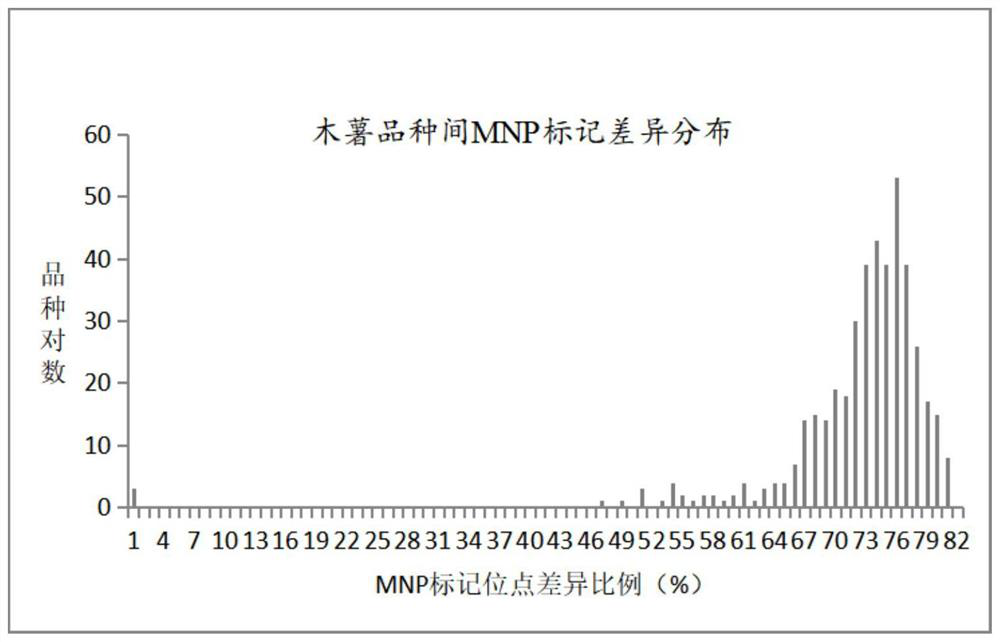
[0079] Taking the analysis of Table 2 in Example 2 as an example, the typing results of 18299 marker sites show that whether it is a repeat experiment or a reproducibility experiment, the results of 623 cassava MNP marker method typing of this invention are accurate. The rate is 99.98%, and the high accuracy shows that in the future application, non-parallel experiments, that is, different time or laboratory test results, have little infl
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap