Sequence Specific and Organism Specific Antimicrobials and Related Materials and Methods
a specific and organism-specific technology, applied in the field of sequence specific and organism-specific antimicrobials and related materials and methods, can solve the problems of antibiotic approval through the u.s. fda, antibiotic development continues to stagnate, and antibiotics represent a relatively poor return on research and development investment,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
[0113]The materials, methods, and embodiments described herein are further defined in the following Examples. Certain embodiments of the present invention are defined in the Examples herein. It should be understood that these Examples, while indicating certain embodiments of the invention, are given by way of illustration only. From the discussion herein and these Examples, one skilled in the art can ascertain the essential characteristics of this invention and without departing from the spirit and scope thereof, can make various changes and modifications of the invention to adapt it to various usages and conditions.
example i
and Methods
[0114]Bacterial Strains and Cell Culture Conditions.
[0115]pAKgfp1 plasmid was obtained from Addgene for TEM-1 β-lactamase resistance gene, bla (43). The plasmid was cloned into chemically competent Zymo DH5a E. coli (Expressys). Untransformed Zymo DH5a was used as a control strain for β-lactamase activity assays. Liquid cultures were grown in 2% LB, incubated at 37° C. and shaken at 225 rpm. Solid cultures were grown on 2% LB broth, 1.5% agar at 37° C. Ampicillin sodium salt (Sigma Aldrich) was used for selection at a concentration of 300 μg / mL for MIC and CFU analysis. Optical density measurements were taken using a Tecan GENios at 562 nm with a bandwidth of 35 nm. All bacterial freezer stocks were stored in 40% glycerol at −80° C.
[0116]MG1655 E. coli cultures were started from individual colonies from 2% LB and 1.5% agar into M9 media. They were diluted 1:10,000 from overnight culture into fresh M9 with respective treatment and grown at 37° C. with 225 rpm shaking. Clini
example ii
the Ribosomal Binding Site and Translation Start Site Reverts β-Lactamase Resistance
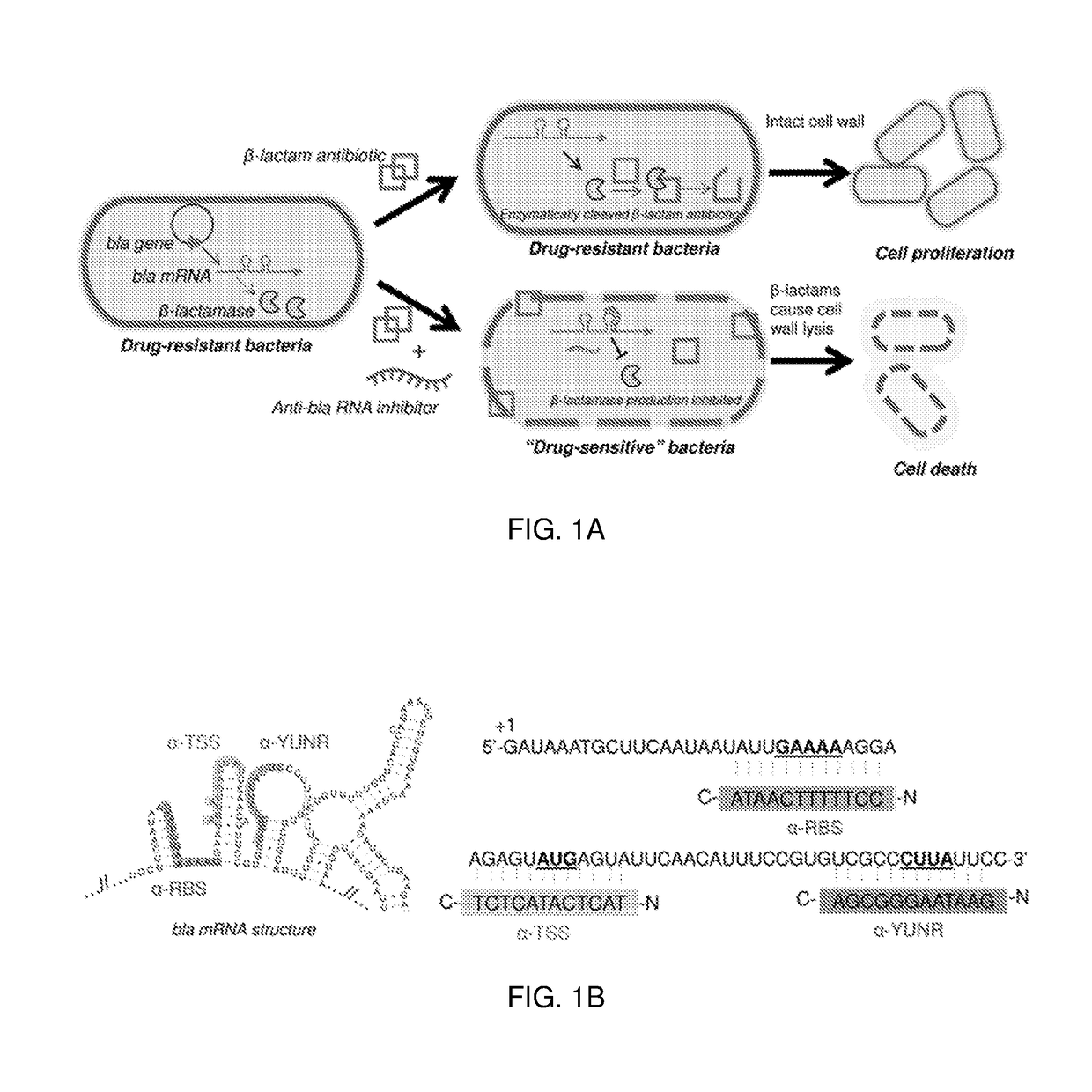
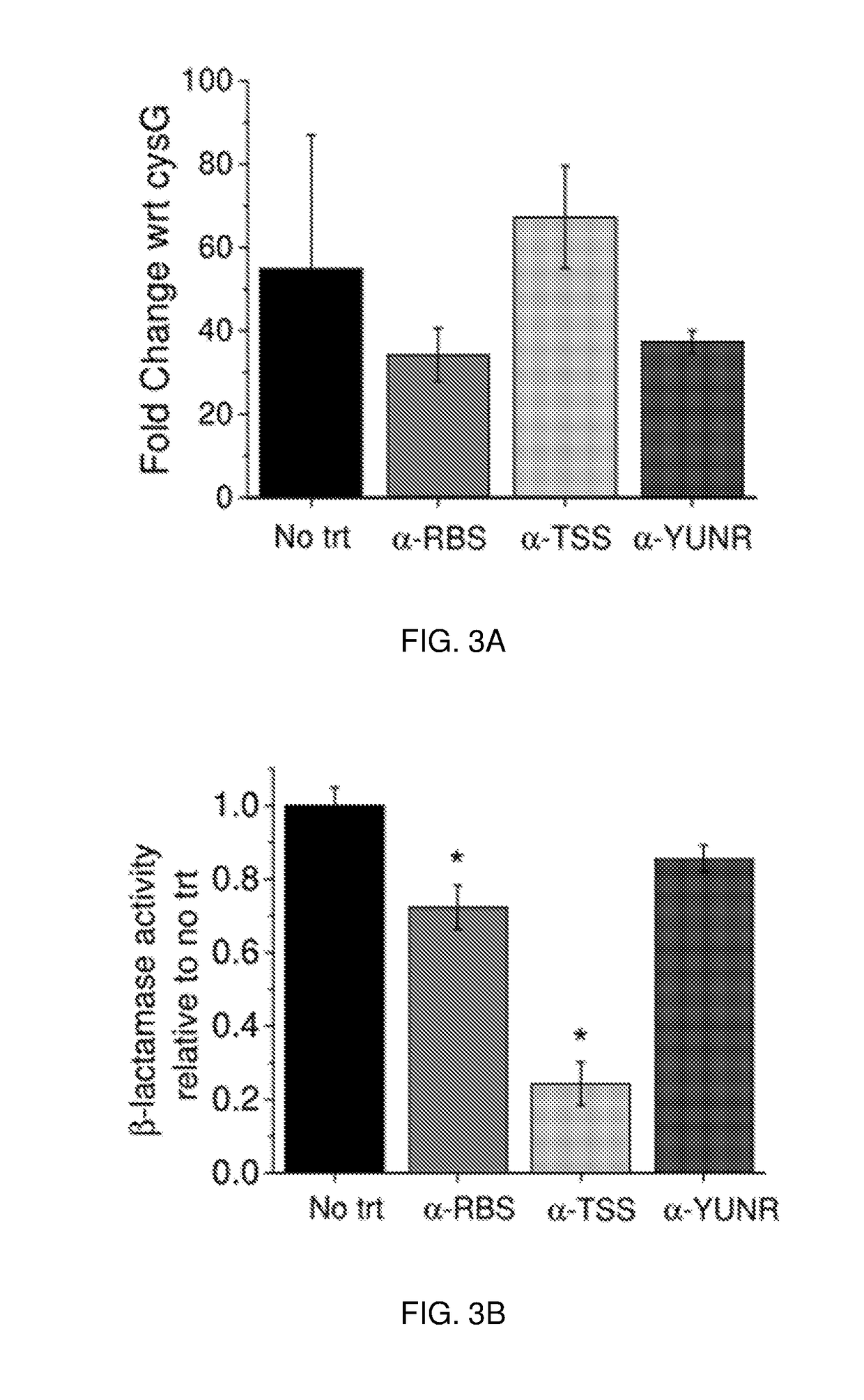
[0137]Protein functional sites on mRNA have been shown to be effective antisense sites for blocking ribosomal binding and migration. These include the ribosomal binding site (RBS) and translation start site (TSS), which are located in the 5′ untranslated region (UTR). The antisense oligomers disclosed and described herein are predicted to sterically hinder the ribosome. In order to prevent the production of truncated, but potentially active, β-lactamase enzyme, the 5′ UTR of bla mRNA was targeted (27).
[0138]Three different antisense oligomers were designed against target sites proximal to the 5′ UTR of bla mRNA: α-RBS; α-TSS; and α-YUNR (FIGS. 1B and 6) (28). Antisense oligomers were designed with the target sequence in the middle of the oligomer, with 3-5 nucleotides flanking the overlapping region (27). To prevent translation of β-lactamase, two short 12-mer antisense oligomers—α-RBS (C-ATAACTTTTTCC-N
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
| Electrical resistance | aaaaa | aaaaa |
| Nucleic acid sequence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap