Method for rapidly detecting mismatching quantity of bases through capillary electrophoresis
A technology of capillary electrophoresis and capillary, applied in the field of biological analysis, to achieve the effect of high repeatability, simple operation, and expanded application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] 1. The capillary adopts a quartz capillary with an inner diameter of 75 μm, a total length of 60 cm, and an effective length of 35 cm. The sample to be tested and the buffer solution flow in the capillary through an 18kV high-voltage power supply.
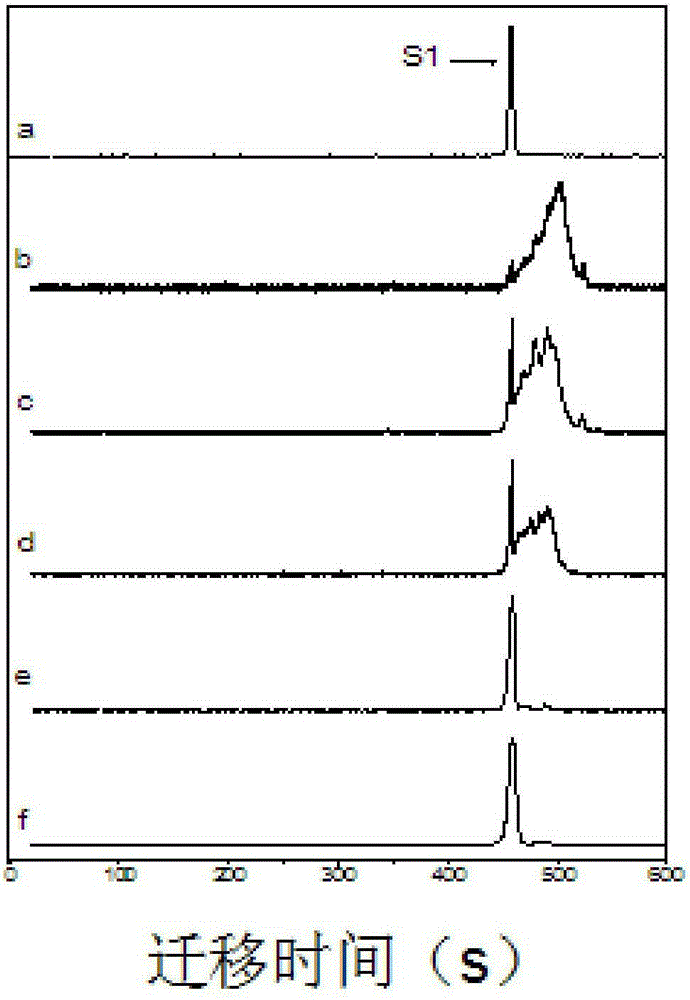
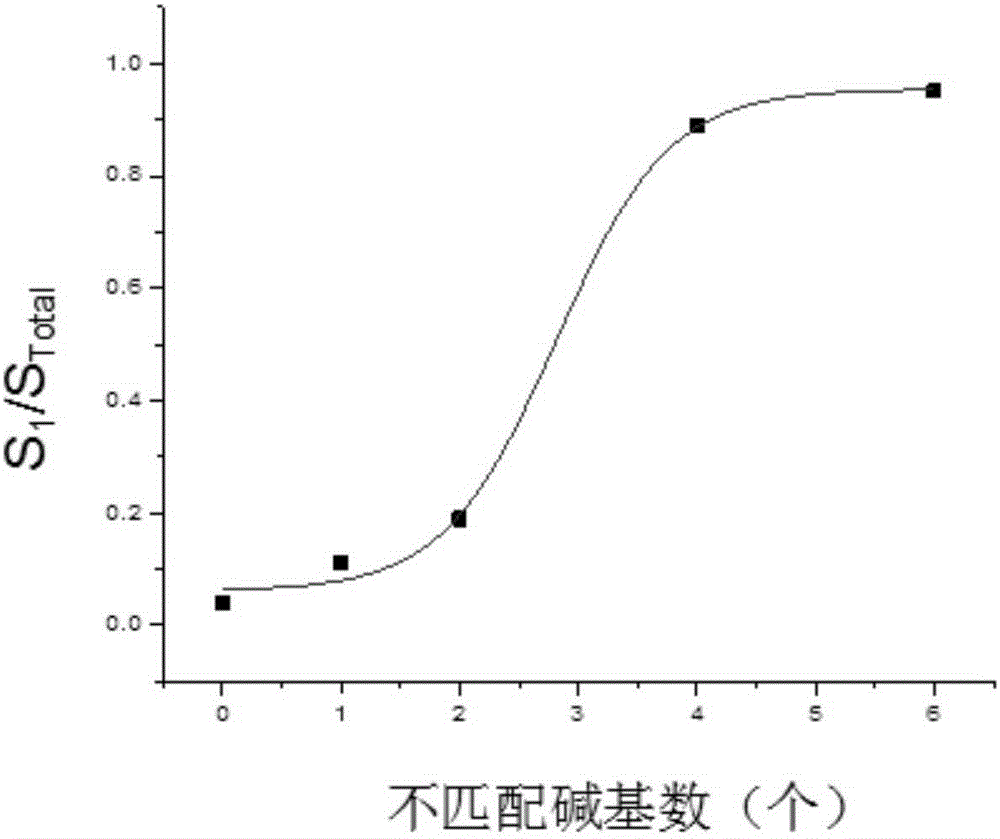
[0020] 2. (1) Design DNA1-FAM fluorescent probe, which includes two parts of fluorescent dye 6-FAM and DNA1. DNA1 is 5'-GGTTGGTGTGGTTGG-3', and the 3' end is coupled with a fluorescent dye; (2) Design five complementary DNAs, which are 5'-CCAACCACACCAACC-3' (zero mismatched bases), 5'-CCAACCACACCAAC G -3' (one mismatch base), 5'-CCAACCACACCAA GG -3' (two mismatched bases), 5'-CCAACCACACC GGGG -3' (four mismatched bases), 5'-CCAACCACA GGGGGG -3' (six mismatched bases); (3) DNA1-FAM and complementary DNA containing different numbers (0, 1, 2, 4, 6) of mismatched bases at the same concentration ratio 2:2, with an interval of 20s in the capillary, inject samples for 20s respectively, and detect by fluorescence capillary elect
Embodiment 2
[0023] 1-2 steps are the same as in Example 1.
[0024] 3. DNA3 to be detected, the sequence is 5'-CCAA GG ACACCAACC-3', carry out capillary electrophoresis, get S 1 / S Total is 0.2, compared with the standard curve, the number of mismatches detected is 2.
PUM
| Property | Measurement | Unit |
|---|---|---|
| The inside diameter of | aaaaa | aaaaa |
| Full length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap