Primer combination and kit for identifying African swine fever virus gene deletion strain and African swine fever epidemic strain through centrifugal microfluidic chip
An African swine fever virus and microfluidic chip technology, applied in the fields of diagnosis and nucleic acid detection, can solve the problems of high-priced detection equipment detection cycle, inability to popularize and use, single detection target, etc., so as to reduce labor and equipment costs and shorten the detection cycle. , the effect of broad market prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1 Structure of microfluidic chips
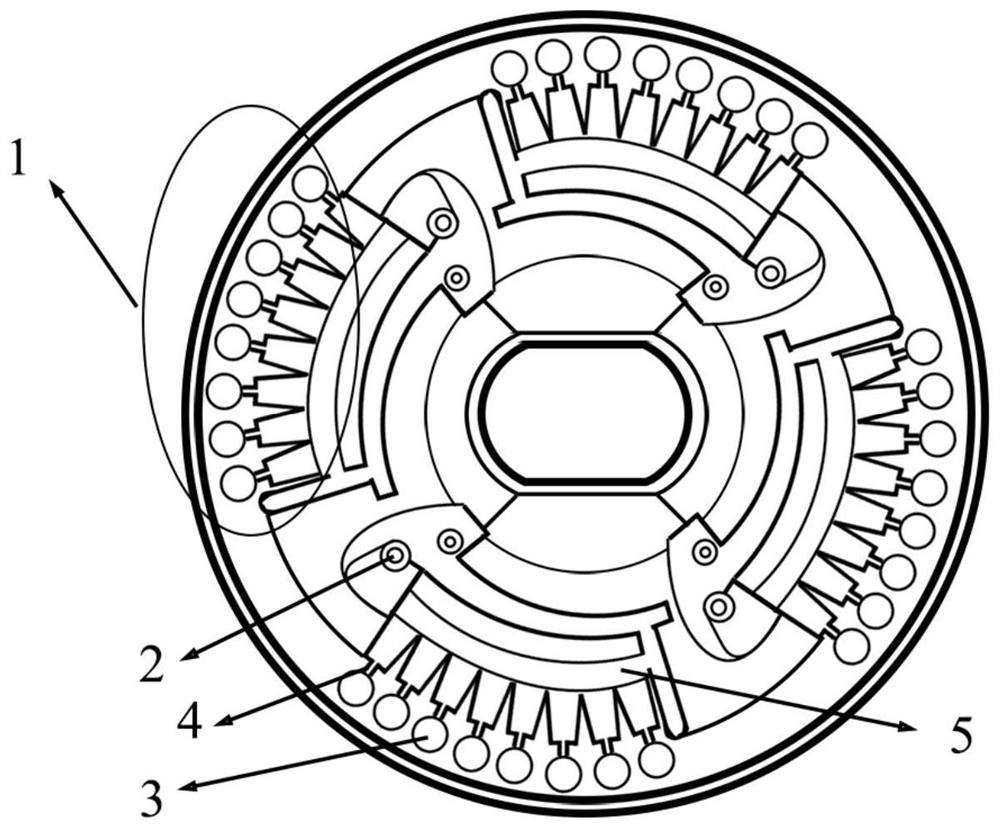
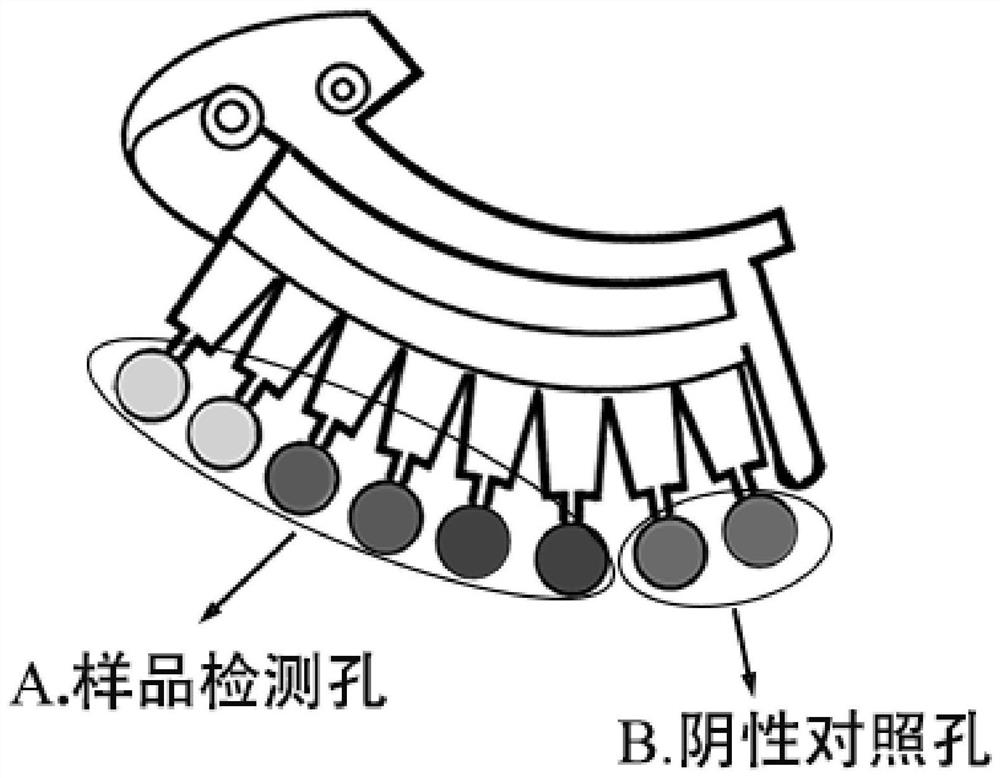
[0055] like figure 1 As shown, the microfluidic chip used in the present invention is a disc-shaped microfluidic chip, the chip is produced by Shanghai Express Diagnostic Products Co., Ltd., has a model of 4 × 8, which includes 4 reaction detection zones 1, Each reaction detection zone 1 includes inventive pool 2, a dispensing pool 5, a capillary micro-valve 4, an amplified pool 3, and the injective pool 2 communicates with the dispensing pool 5, in the injective pool 2 There is a sample hole, and the dispensing pool 5 communicates with the waste liquid pool and exhaust hole through a curved channel; each reaction detection zone is provided with 8 amplification pools.
[0056] The main function of the invasive pool 2 is loaded reaction liquid; the main function of the distribution pool 5 is to uniformly distribute the reaction liquid to the amplification pool; the amplification pool 3 specifically realizes the LAMP reaction; the capi
Embodiment 2
[0057] Example 2 primer screening
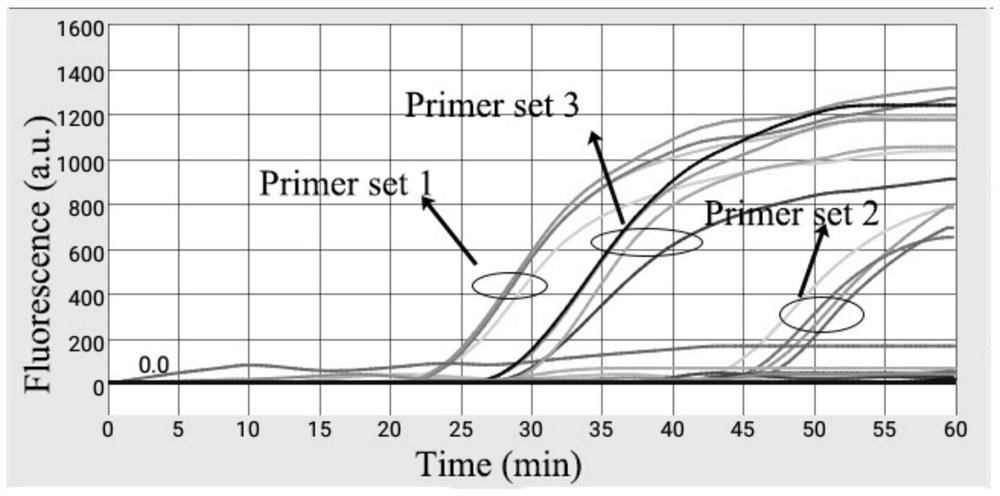
[0058] Retrieves all ASFV full-length genome sequences from the GenBank database, and the homologous analysis of the BLAST software is found to find a target gene sequence corresponding to the viral genome relative to the viral genome. Depending on the resulting target sequence, the LAMP primer is designed using PrimeRexplorer V5 and synthesized.
[0059] After the synthesized primer is dissolved, the primer screening is performed, and the screening process is as follows:
[0060] 1, ASFV-MG505-2R primer screening
[0061] 3 groups of primers designed for ASFV-MG505-2R gene fragments, primer sequences are as follows:
[0062] First group:
[0063] MG505-2R-F3-1: agggtgatcaattructg;
[0064] MG505-2R-B3-1: TCTTCTTGGTATTTTGGAGAA;
[0065] MG505-2R-FIP-1:
[0066] Cgcgtcttgaatcaatggtaagaatccataagtatgaaaaccaaatcg;
[0067] MG505-2R-BIP-1: atacgttgaaaaatgccccccagcatgtgttttgtagca.
[0068] Second Group:
[0069] MG505-2R-F3-2: ccaaaaataccaagaagagct;
[0
Embodiment 3
[0152] Example 3 composition of the kit
[0153] The kit according to the present invention, comprising the microfluidic chip described in Example 1, the primer group and three corresponding African swine fever virus gene deletion strains and African swine fever epidemic strains Lamp method positive standards also include a constant temperature amplification premix.
[0154] The primer group sequences used to identify the deletion strains of African swine fever virus genes and African swine fever include primers for identifying diagnosis of ASFV-MG505-2R (hereinafter can also be referred to as ASFV-MGF) for identifying diagnostic ASFVs. -CD2V primers for identifying primers for diagnosing ASFV-P72. 5 primers in each group were an outer primer F3 / B3: an inner primer FIP / BIP: The ring LB / LF was 1: 8: 4.
[0155] Through gene synthesis, cloning and connecting T-load, transformation, challenge, racemic, sequencing identification, respectively, the correct three kinds of LAMP
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap