Plant vernalization independence (VIP) genes, proteins, and methods of use
a technology of plant genes and proteins, applied in the field of plant genes involved in regulating flowering, can solve the problems of increasing and restricting the use of many traditionally utilized chemical compounds, and achieve the effects of reducing flc rna expression, increasing or decreasing activity, and reducing production costs and labor requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Experimental Procedures: VIP4 Gene
[0302]This example describes the experimental procedures used to identify, clone, and characterize the VIP4 gene.
Plant Material and Growth Conditions
[0303]Introgression line Col:FRISF2 consists of the FRI locus from ecotype San Feliu-2 (FRISF2) introgressed into the Columbia (Col) ecotype through six successive backcrosses and made homozygous by self-pollination (Lee et al. (1994) Plant J. 6, 903-909). Line FN231 contains a fast-neutronflc allele isolated in the Col:FRISF2 background, and is identical with flc-1 described by Michaels and Amasino ((1999) Plant Cell, 11, 949-956). Line FN235, containing a fast-neutronfri allele isolated in the Col:FRISF2 background, is as described by Michaels and Amasino ((1999) Plant Cell, 11, 949-956). The ld-1 mutant in the Col background was obtained from the Arabidopsis Biological Resource Center (ABRC) at The Ohio State University. Standard growth conditions were 22° C. under 100-180 μmol m-2·sec-1 of cool white f
example 2
VIP4 Gene: Identification, Cloning, and Characterization
[0308]This example describes the identification, cloning, and characterization of the VIP4 gene.
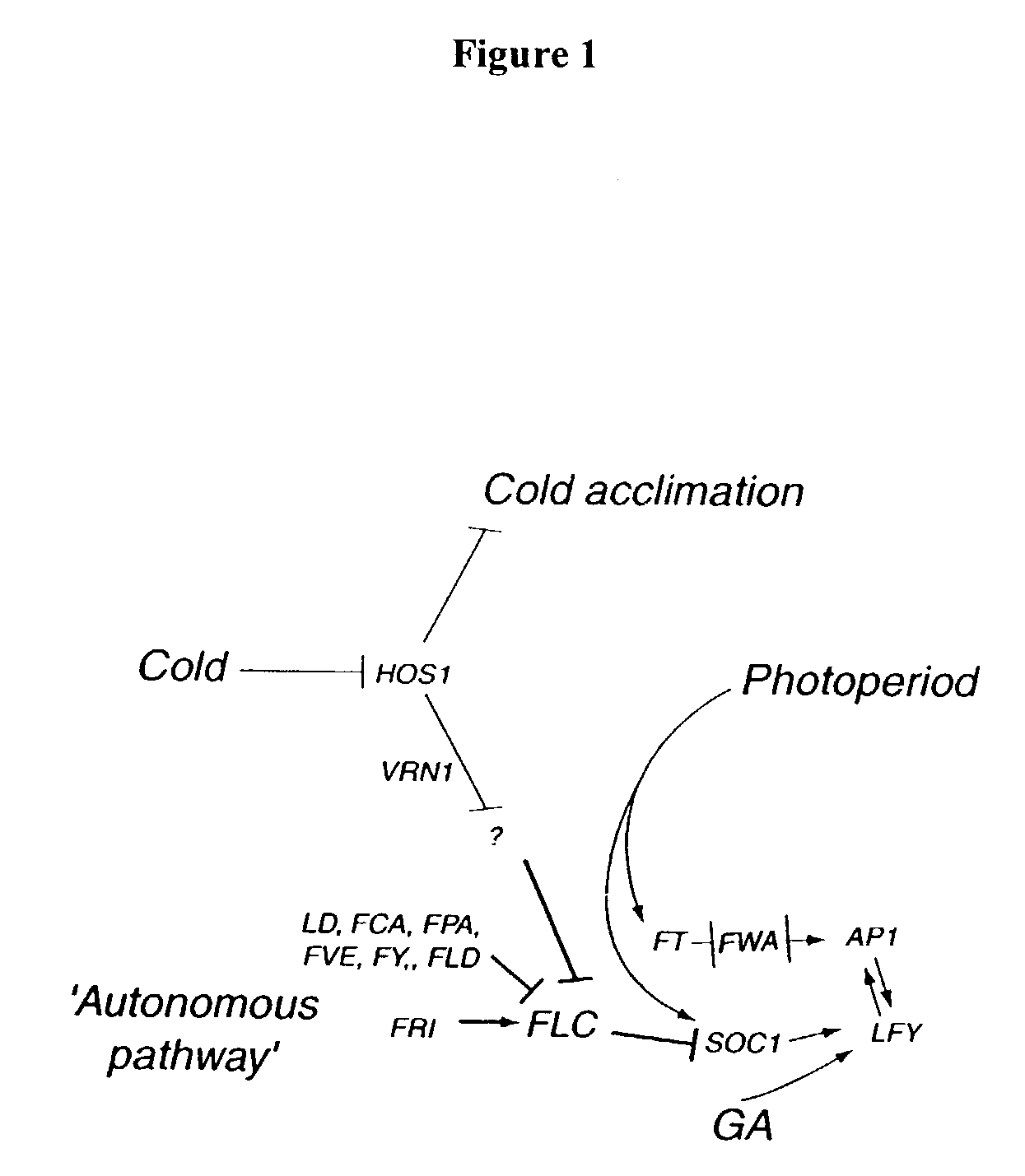
A Genetic Screen for Activators of FLC
[0309]To identify potential activators of FLC, the winter-annual, Col:FRISF2 genetic background, was mutagenized and screened for recessive mutations that conferred cold-independent, early flowering. Early-flowering lines were rescreened by assaying for reduced FLC RNA expression in seedlings, where FLC RNA is typically easily detectable. To eliminate further consideration of lines with mutations in either the FLC or the FRI genes, mutants were also used in genetic complementation analysis with lines FN231 and FN235, carrying loss-of-function mutations in the FLC and FRI genes, respectively. Early-flowering lines that exhibited reduced FLC RNA expression, and that were not likely to represent new alleles of FLC or FRI, were sorted into allelic groups through complementation analysis. This strategy r
example 3
Experimental Procedures: VIP3 Gene
[0319]This example describes the experimental procedures used to identify, clone, and characterize the VIP3 gene.
Growth Conditions.
[0320]Arabidopsis seeds were either planted directly into artificial soil mix (Baccto Hi-Porosity Mix, Michigan Peat Co., Houston, Tex.) or surface-sterilized and germinated on sterile Germination Medium [2.5 mM 2-[N-Morpholino]ethanesulfonic acid, 375 mg / l Peters 20-20-20 Fertilizer (Scotts Company, Marysville, Ohio), pH adjusted to 5.7 with KOH, supplemented with 0.8% (w / v) Phytagar (Life Technologies, Gaithersburg, Md.)]. Standard growth conditions were 60-100 μmoles m−2 s−1 of fluorescent lighting in a 16h-light / 8h-dark photoperiod at 22° C. and ˜50% relative humidity. For a vernalizing cold treatment, seeds on Germination Medium were first placed at 4° C. under 20-50 μmoles m−2 s−1 of fluorescent lighting in a 8h-light / 16h-dark photoperiod for 30 or 70 d. To evaluate flowering time, individual plants were grown
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap