Recombinant host cells comprising phosphoketolases
A technology for recombining host cells and phosphoketolase, which can be used in recombinant DNA technology, enzymes, bacteria, etc., and can solve problems such as cost increase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
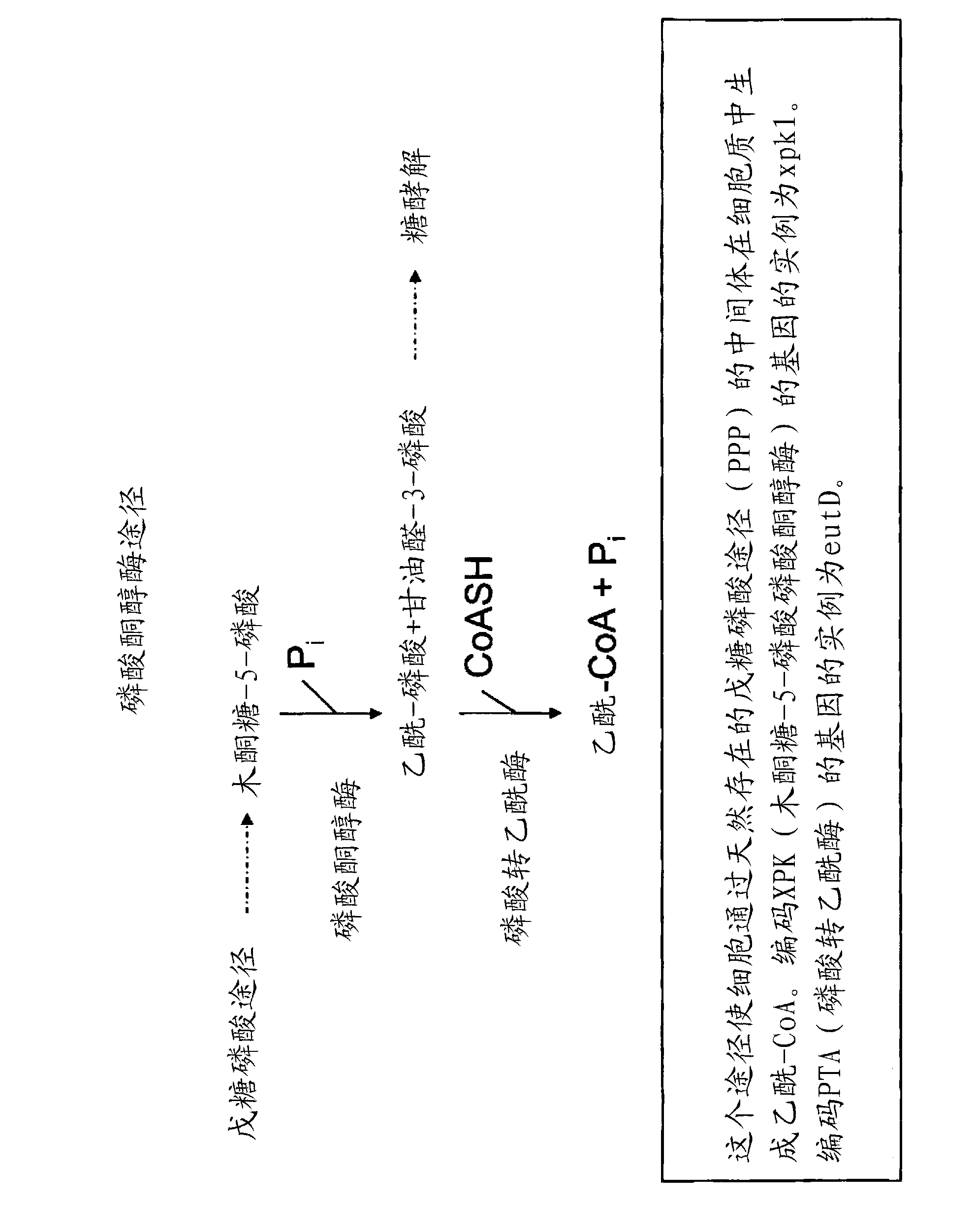
[0388] Construction of phosphoketolase / phosphotransacetylase expression cassette
[0389] The xpk1 and eutD genes (GenBank GI numbers are 28379168 (SEQ ID NO: 172) and 28377658 (SEQ ID NO: 1111) respectively) were obtained using primers N1039 and N1040 (for xpkl) and N1041 and N1042 (for eutD) Obtained from Lactobacillus plantarum (ATCC No.BAA-793) by polymerase chain reaction. The primer sequences of N1039, N1040, N1041 and N1042 respectively correspond to SEQ ID NO:639-642.
[0390] The xpk1 and eutD genes were obtained from PacI digested pRS423::CUP1-alsS+FBA-budA by overlapping polymerase chain reaction with yeast terminators containing opposite (CYC and ADH terminators, described in US Patent Application Publication 20090155870, incorporated herein by reference) DNA fragment fusion (Yu et al., Fungal Genet. Biol. 41:973-981, 2003). The resulting PCR product was cloned into an E. coli yeast shuttle vector using the gap repair method (Ma et al., Genetics 58:201-216; 1981
Embodiment 2
[0392] Construction of phosphoketolase / phosphotransacetylase integration vector
[0393] The expression cassette (GPD-xpk1+ADH1-eutD) of the pRS426::GPD-xpk1+ADH1-eutD vector was prepared by restriction enzyme digestion with EcoRI and SacI. The resulting expression cassette was ligated into the yeast integration vector pUC19-URA3-MCS, which was also prepared by EcoRI and SacI restriction enzyme digestion.
[0394] The vector pUC19-URA3MCS is based on pUC19 and contains the sequence from the URA3 gene of Saccaromyces cerevisiae located within the multiple cloning site (MCS). pUC19 (American Type Culture Collection, Manassas, VA; ATCC #37254) contains the pMB1 replicon and the gene encoding β-lactamase for replication and selection in E. coli. In addition to the coding sequence of URA3, the upstream to downstream sequences of this gene were included for the expression of URA3 in yeast. The vectors are useful for cloning purposes and as yeast integration vectors.
[0395] The
Embodiment 3
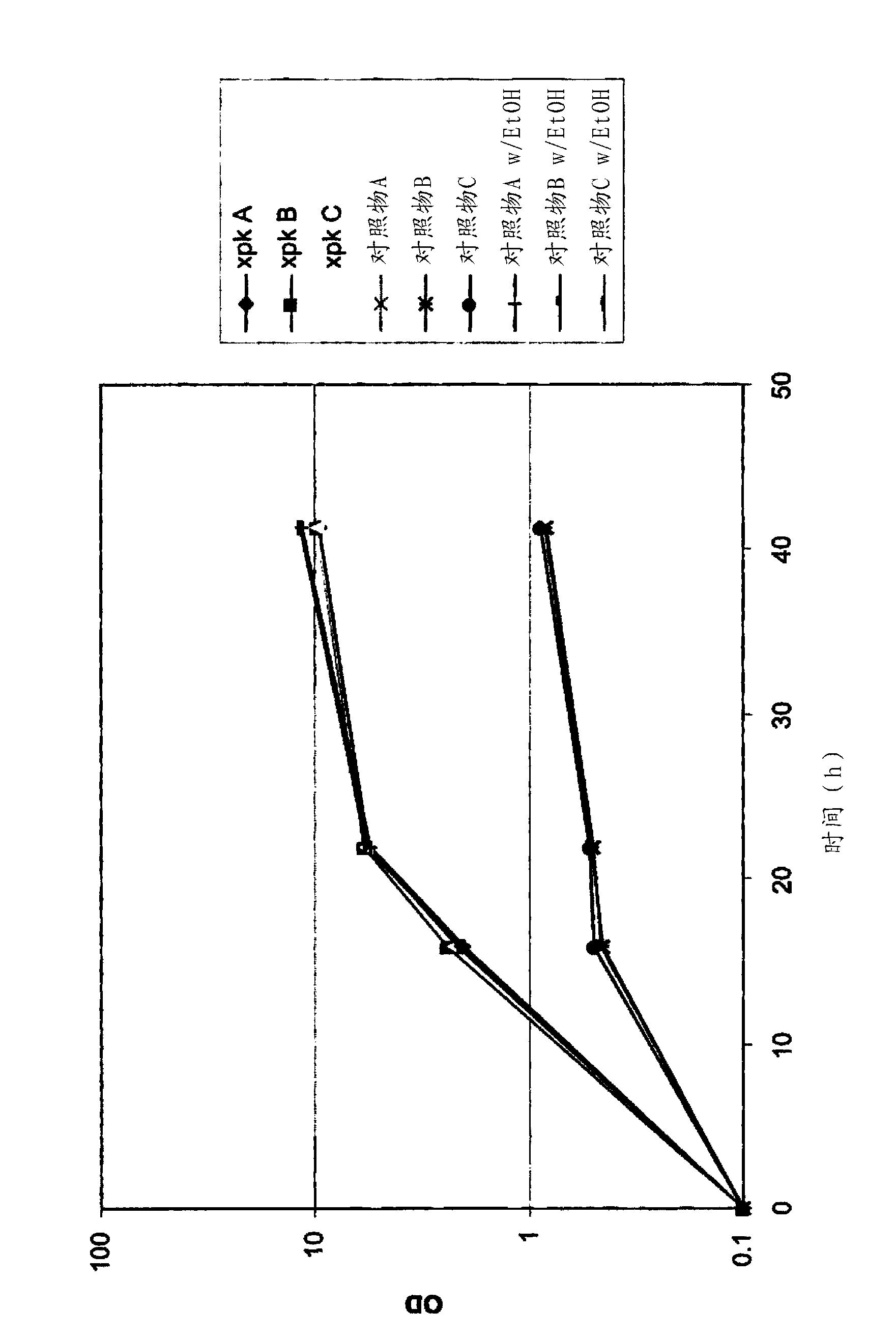
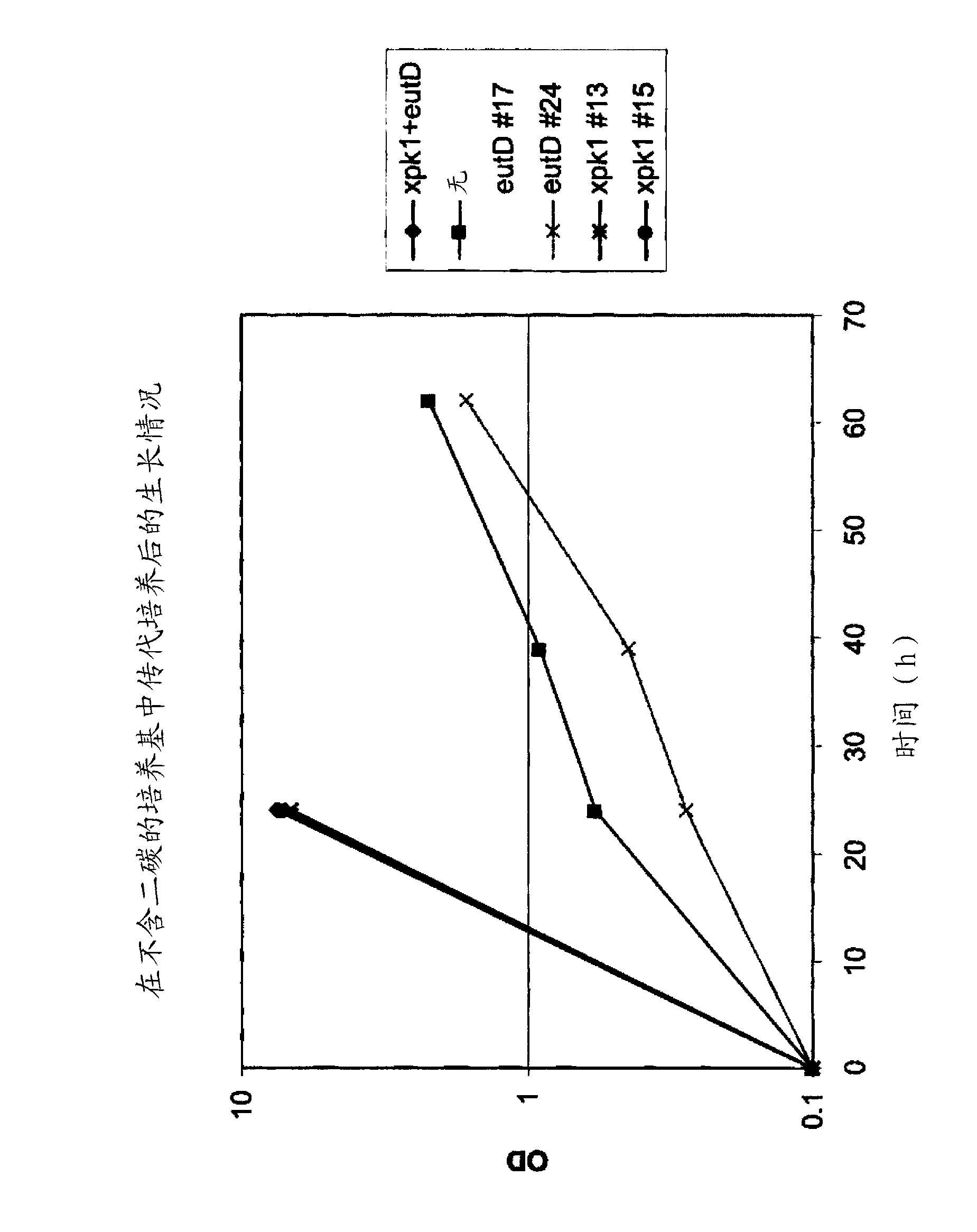
[0400] Construction of pyruvate decarboxylase knockout (PDC- KO) yeast strain
[0401] Control and phosphoketolase pathway vectors as described in Example 2 were linearized with Aflll restriction enzyme and transformed into strain BP913 (CEN.PK113-7DΔura3::loxPΔhis3Δpdc6Δpdc1::ilvD(Sm)Δpdc5::sadB) to Control and phosphoketolase pathway strains were generated. Strain BP913 is further described in Example 10.
[0402] Transformed cells were plated on synthetic flex media without uracil, containing ethanol as a single carbon source (1% v / v), and passed through the Screening was performed by PCR to confirm integration at the pdc1 locus. Integration at the Δpdc1::ilvD(Sm) locus results in loss of ilvD(Sm).
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap