Method of simply, conveniently and quickly measuring protease activity
A protease and enzyme activity technology, applied in the field of bioengineering, can solve the problems of large sample demand and long time required, and achieve the effect of small sample demand, short time consumption and simple process operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
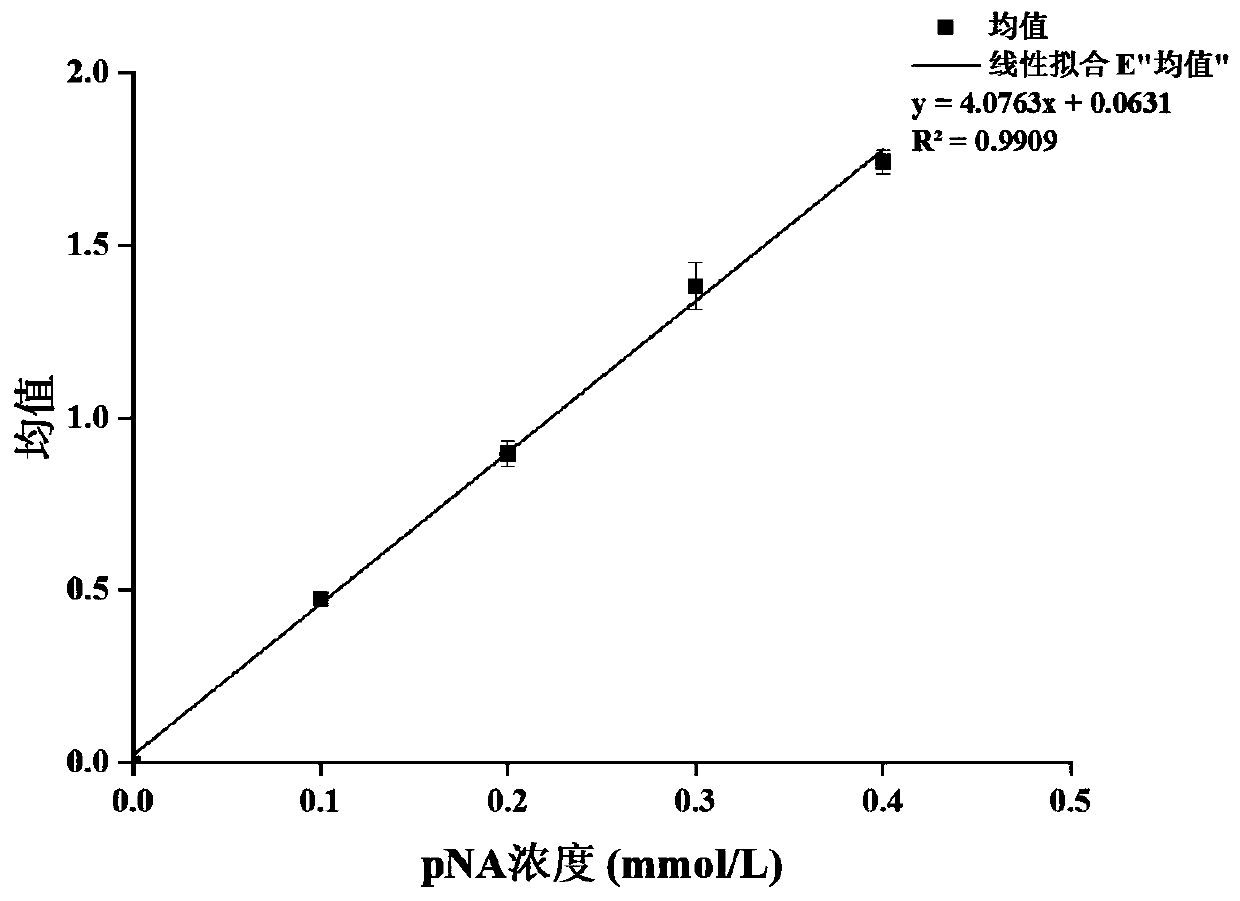
[0028] Embodiment 1: AAPY substrate assays the drafting of the standard curve of protease enzymatic activity
[0029] 1) Prepare different concentrations of p-nitroaniline solutions with DMSO.
[0030] 2) Composition of the reaction system: 80 μL of phosphate buffer (pH=7.5), 20 μL of p-nitroaniline solution, and 100 μL of phosphate buffer (pH=7.5).
[0031] 3) Take tube 0 without p-nitroaniline as blank, at OD 410 The absorbance was measured respectively.
[0032] 4) Take the absorbance A as the ordinate, and the final concentration C of p-nitroaniline as the abscissa, draw a standard curve (the curve should pass through the zero point). Calculate the concentration of p-nitroaniline when the absorbance is 1 according to the drawing, which is the absorbance constant K value. standard curve as figure 1 shown.
Embodiment 2
[0033] Embodiment 2: Utilize AAPY substrate to measure the enzymatic activity of various proteases
[0034] 1. Using AAPY substrate to measure the enzyme activity of 1398 neutral protease
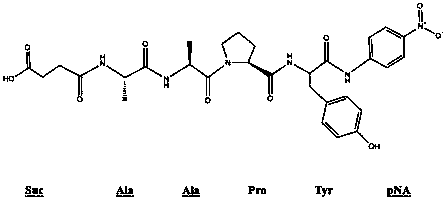
[0035] 1) Prepare 4mM AAPY substrate solution with DMSO;
[0036] 2) Reaction system composition: 80 μL phosphate buffer (pH=7.5), 20 μL AAPY substrate solution, 100 μL diluted enzyme solution (3-4 U / mL) of the sample to be tested;
[0037] 3) Mix well, let stand at 40°C for 10 minutes, measure OD 410 Absorbance value at time;
[0038] 4) Calculate the enzyme activity in the enzyme solution to be tested, and express the result with one decimal place.
[0039] 2. Determination of enzyme activity of alkaline protease derived from Bacillus clausii using AAPY substrate
[0040] 1) Prepare 4mM AAPY substrate solution with DMSO;
[0041] 2) Reaction system composition: 80 μL borax-sodium hydroxide buffer solution (pH=10.5), 20 μL AAPY substrate solution, 100 μL enzyme solution (3-4 U / mL) of
Embodiment 3
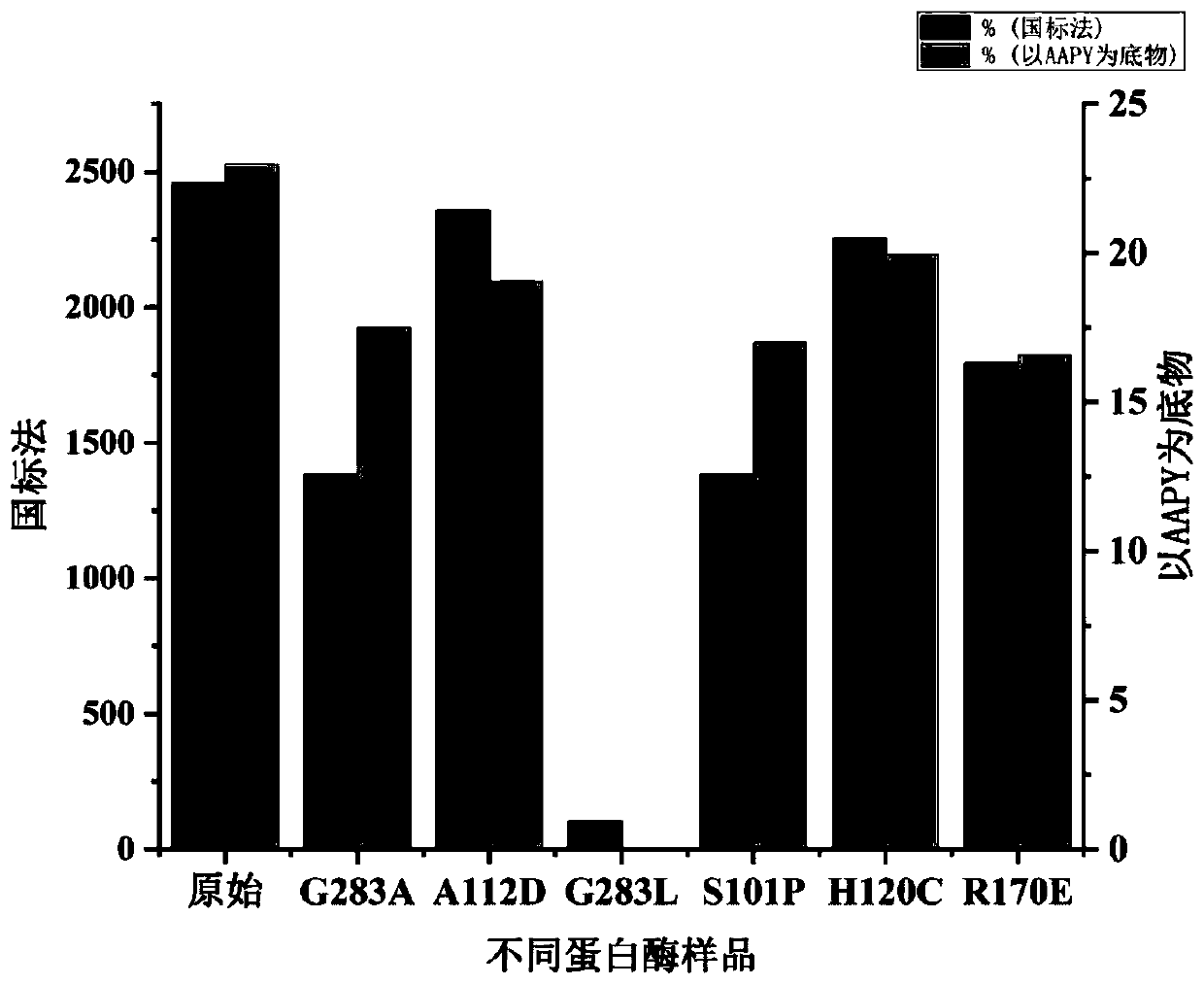
[0061] Embodiment 3: Utilize national standard method and method of the present invention to measure a plurality of alkaline protease samples simultaneously
[0062] The alkaline protease gene and its variants derived from Bacillus clausii were expressed in Bacillus subtilis to obtain multiple strains, and the enzyme activity data of these strains were determined by fermentation in this experiment. The protease gene number derived from Bacillus clausii is: CP019985.1. The original strain and 5 mutant strains were selected for this experiment. Naming rules for mutant strains: "amino acid replaced by the original amino acid position" is used to represent the mutant strain, and the amino acid residue uses the single-letter symbol form in the recognized nomenclature, such as G283A, indicating that the amino acid at position 283 is derived from glycine (G) of the parent protease Replaced by alanine (A), the numbering of the positions corresponds to the amino acid sequence numbering o
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap