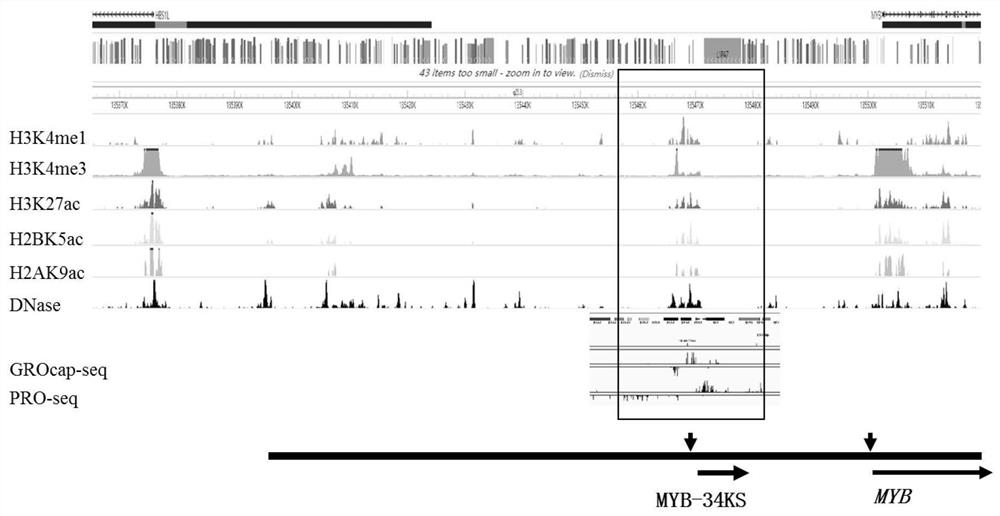
IncRNA transcribed in upstream 34kb region of MYB gene, expression vector and application thereof
An upstream and regional technology, applied in the direction of DNA/RNA fragment, recombinant DNA technology, and the determination/inspection of microorganisms, which can solve the problems of unclear effect and impact research.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Example Embodiment
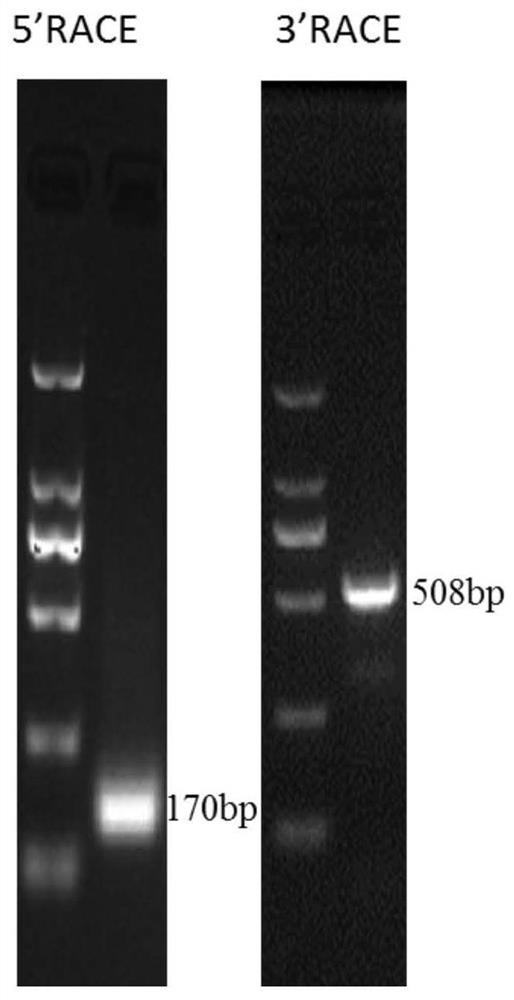
[0054] Example 1 Cloning of the full-length sequence lncRNA MYB-34KS embodiment
[0055] Get the RACE technique lncRNA MYB-34KS cDNA full-length sequence, amplified by RACE unknown sequence ends and then the next step after the amplification, sequencing specific sequence.
[0056] Methods as below:
[0057] (1) Total RNA extraction
[0058] K562 cell total RNA extraction using Trizol method.
[0059] (2) RACE First strand cDNA synthesis
[0060] This step is mainly provided by the company Takara RACE 5 ' / 3'Kit kit complete.
[0061] 3'-RACE-Ready cDNA and 5'-RACE-Ready cDNA generated by doubled sterilized H 2 O diluted in at -20 ℃.
[0062] (3) RACE-PCR reactions
[0063] A first round of RACE-PCR reactions, respectively, using the UPM primer and the 5'-end or 3'-end universal gene-specific primer (gene-specific primer, GSP) for PCR amplification.
[0064] lncRNA MYB-34KS PCR primer sequences:
[0065] 5'RACE-MYB-34KS-GSP: 5'-accactgttaacccaattgacgcctgact-3 ';
[0066] 3'RACE-MYB
Example Embodiment
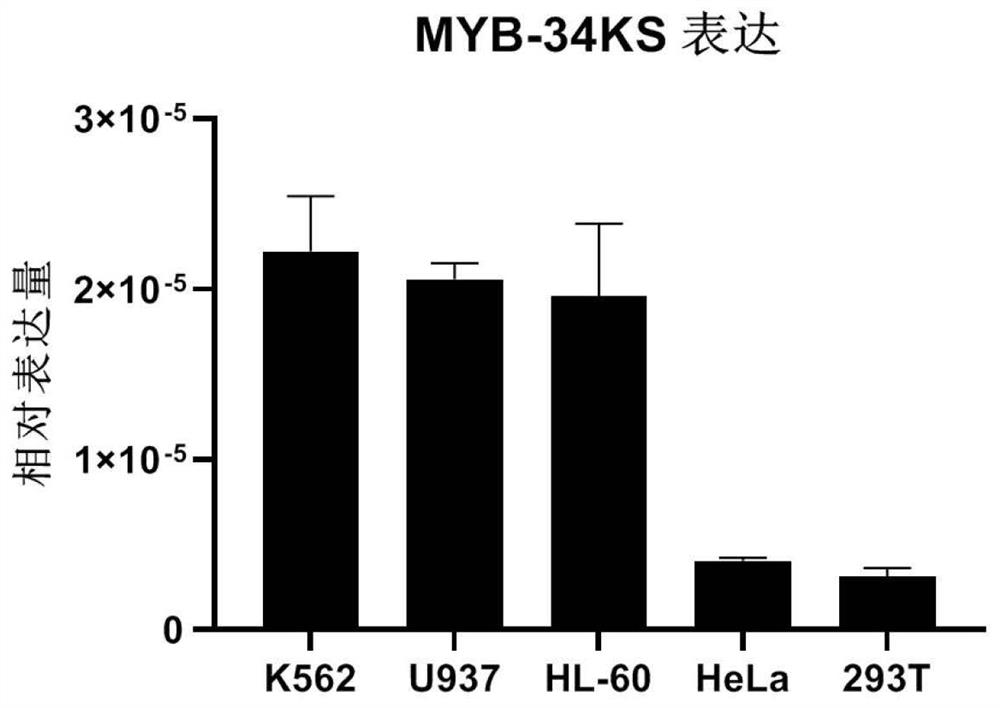
[0102] Example 2 lncRNA MYB-34KS expression level detected in leukemia cell lines embodiment
[0103] (1) Cell Culture
[0104] K562 human erythroleukemia cells, U937 human acute myeloid leukemia cells, human promyelocytic acute leukemia HL60 cells, of Hela cells and 293T cells were purchased from ATCC (American Type Culture Collection). K562, U937, HL60 cells were treated with RPMI 1640basic (Gibco, # C11875500BT) culture medium, added to a final concentration of 10% fetal calf serum (Gibco, # 10099-141C) and a final concentration of 1% penicillin and streptomycin (HyClone, # SV30010). Hela 293T cells and medium plus 10% fetal calf serum and 1% penicillin and streptomycin and cultured in DMEM (Gibco, # C11965500BT). Cells were seeded good temperature was 37 ℃, CO 2 A volume fraction of 5% of the cell culture incubator.
[0105] (2) Extraction of total RNA
[0106] Total cellular RNA was extracted using Trizol each method.
[0107] Cells were cultured in 10cm cell culture dishes, K56
Example Embodiment
[0125] Example 3 lncRNA MYB-34KS overexpression and knockdown vector constructed embodiment
[0126] Example 3 This example, using the full-length sequence lncRNA MYB-34KS obtained from Example 1, over-expression construct and knockdown vector. Example The plasmid was diluted to 1 T 1ng / μL, in order to be subcloned PCR as a template.
[0127] 1, the vector pLVX-MYB-34KS overexpression
[0128] (1) pLVX-IRES-Neo plasmid pLVX-MYB-34KS overexpression vector. The first pLVX-IRES-Neo plasmid and MYB-34KS sequence selected endonuclease restriction site in two, respectively, with EcoRI and XbaI. Then add restriction site designed subcloning primer sequences are as follows:
[0129] MYB-34KS-EcoRI-F: 5'-ggaattcacacactttataattccatattgttccctcatg-3 '
[0130] MYB-34KS-XbaI-R: 5'-gctctagattgtttttgtttttccttccataacatacatttg-3 '
[0131] (2) PCR adding restriction enzyme sites
[0132] reaction system:
[0133]
[0134]
[0135] Reaction conditions: 98 ℃ denaturation 10s, 55 ℃ 5s, 72 ℃ 1min
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap