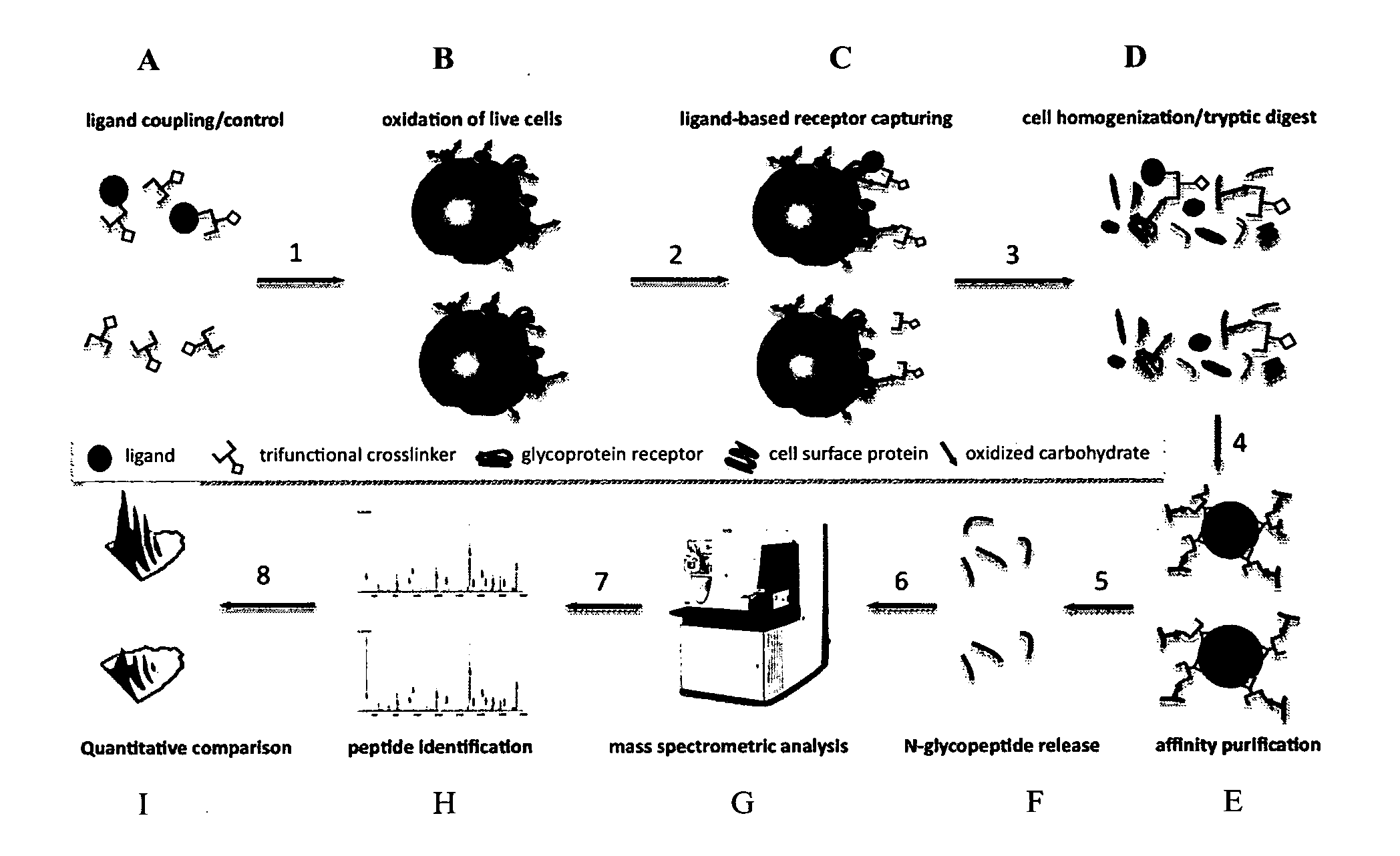
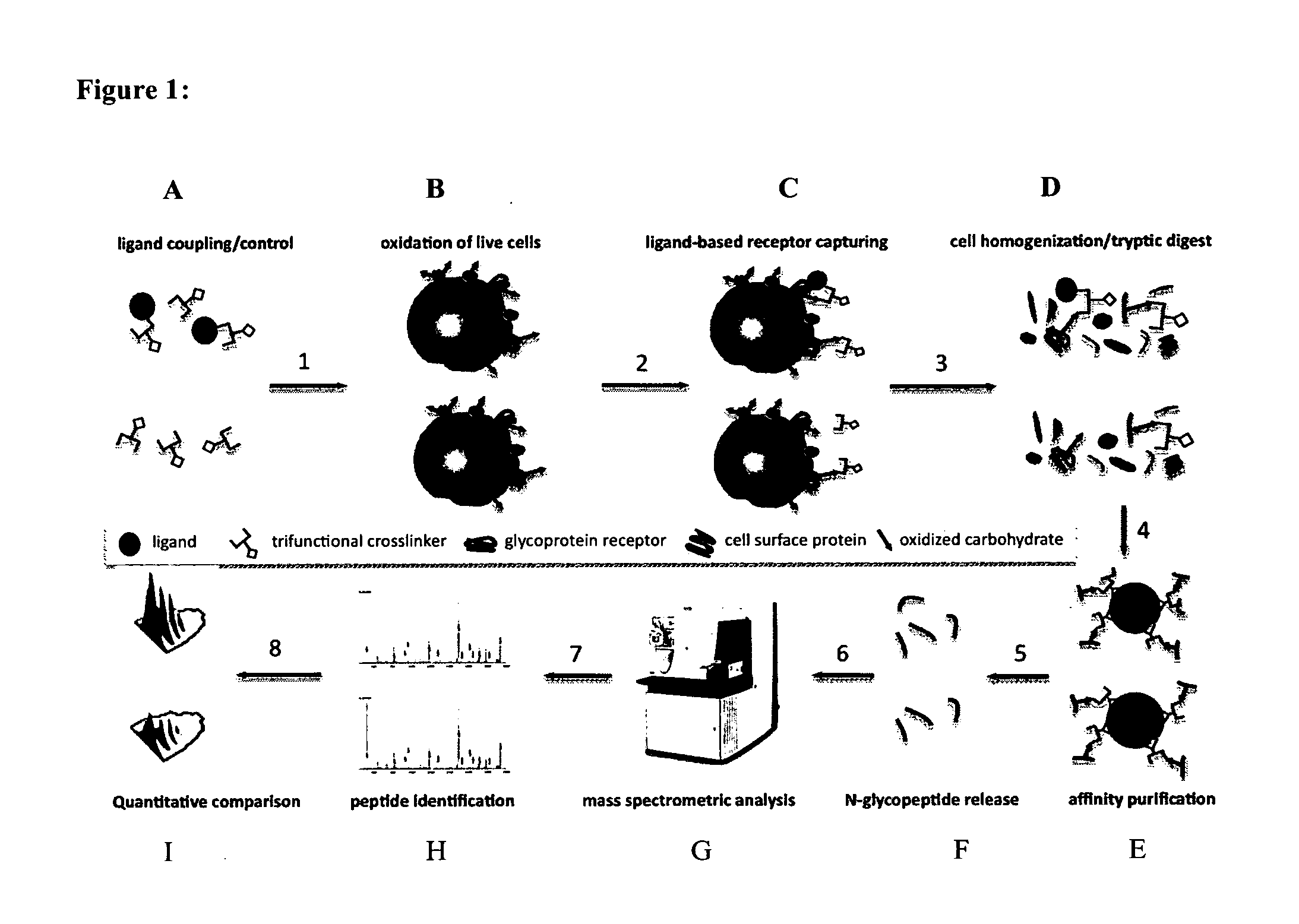
Trifunctional crosslinking reagents
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Synthesis of Crosslinker Joy-06-16
[0212]
(a) Synthesis of (2S)-2-[6-(6-{[(tert-Butoxy)carbonyl]amino}hexanamido)hexanamido]-5-methoxy-5-oxopentanoic Acid (1)
[0213]
[0214]To a solution of (2S)-2-Amino-5-methoxy-5-oxopentanoic acid (9.8 g, 22 mmol; synthesized according to Glenn, M. P. et al, J. Am. Chem. Soc. 2003, 125, 640) in MeOH (130 mL) was added 2,5-Dioxopyrrolidin-1-yl 6-(6-{[(tert-butoxy)carbonyl]amino}hexanamido)hexanoate (5.7 g, 29 mmol; synthesized according to Srinivasan, B. and Huang, X. Chirality 2008, 20, 265) and then to the solution was added TEA (9.4 mL, 67 mmol). After stirring for 30 min at room temperature, the reaction mixture was concentrated under reduced pressure and dissolved in EtOAc (200 mL) and then washed with 1N HCl (100 mL) and washed with brine and dried over MgSO4 and concentrated under reduced pressure and purified by flash chromatography (CH2Cl2:MeOH=10:1 to CHCl3:MeOH:H2O=85:15:1 to CHCl3:MeOH:H2O=65:25:4) providing the desired compound as a white foam
example 2
Synthesis of Crosslinker Joy-05-125
[0237]
(a) Synthesis of tert-Butyl N-[(5S)-5-[(3-{2-[2-({9-[(3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4-d]imidazolidin-4-yl]-5-oxononyl}oxy)ethoxy]ethoxy}propyl)carbamoyl]-5-{[(9H-fluoren-9-ylmethoxy)carbonyl]amino}pentyl]carbamate (9)
[0238]
[0239]Fmoc-N-ε-Boc-L-Lysine (1.7 g, 3.7 mmol) was dissolved in DMF (20 mL) and then to the solution were added DIPEA (0.63 mL, 3.7 mmol) and HBTU (1.7 g, 4.4 mmol) and after 10 min then to the reaction mixture was added a solution of 1-N-biotinyl-4,7,10-trioxatridecane-1,13-diamine (1.8 g, 4.1 mmol) in DMF (5 mL) and stirred at room temperature for 1 hr and then the solvent was evaporated under reduced pressure and purified by flash chromatography (CHCl3:MeOH:H2O=10:6:1) providing the desired compound as a white viscous foam (2.6 g, 78%).
[0240]TLC (CH2Cl2:MeOH, 10:1 v / v): RF=0.2; 1H-NMR (400 MHz, CD3OD): δ 7.86 (d, J=7.5 Hz, 2H), 7.73 (dd, J=6.5, 4.7 Hz, 2H), 7.48-7.36 (m, 4H), 4.53 (dd, J=7.8, 4.3 Hz, 1H), 4.46 (t
example 3
Ligand-Based Receptor Capturing with Insulin
[0264](a) Ligand Coupling to Trifunctional Cross-Linker Joy-05-125 (Obtained from Example 2)
[0265]50 μg Joy-05-125 (100 mM in DMSO) was added to 100 μg of insulin (I9278, Sigma-Aldrich) in 10 μl HEPES pH8.2 to obtain a ratio of cross-linker:ligand of approximately 2:1. For the control sample, 50 μg Joy-05-125 (100 mM in DMSO) was added to a quenching solution (10 mM Glycine in 10 μl HEPES pH8.2). Reactions were carried out for approximately 1 h at room temperature.
(b) Harvesting of Cells and Oxidation of Cell Surface Glycoproteins.
[0266]2×108 cells (Jurkat T) were collected in a 50 ml tube and washed with phosphate buffered saline (PBS, pH7.4). Subsequently, cells were oxidized for 15 min in the dark at 4° C. with 1.5 mM sodium-meta-periodate (Thermo Scientific) in labeling buffer (PBS, pH6.5). The cell pellet was washed once with 50 ml labeling buffer to remove most of the sodium-meta-periodate and to deplete dead cells / fragments.
(c) Ligan
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap