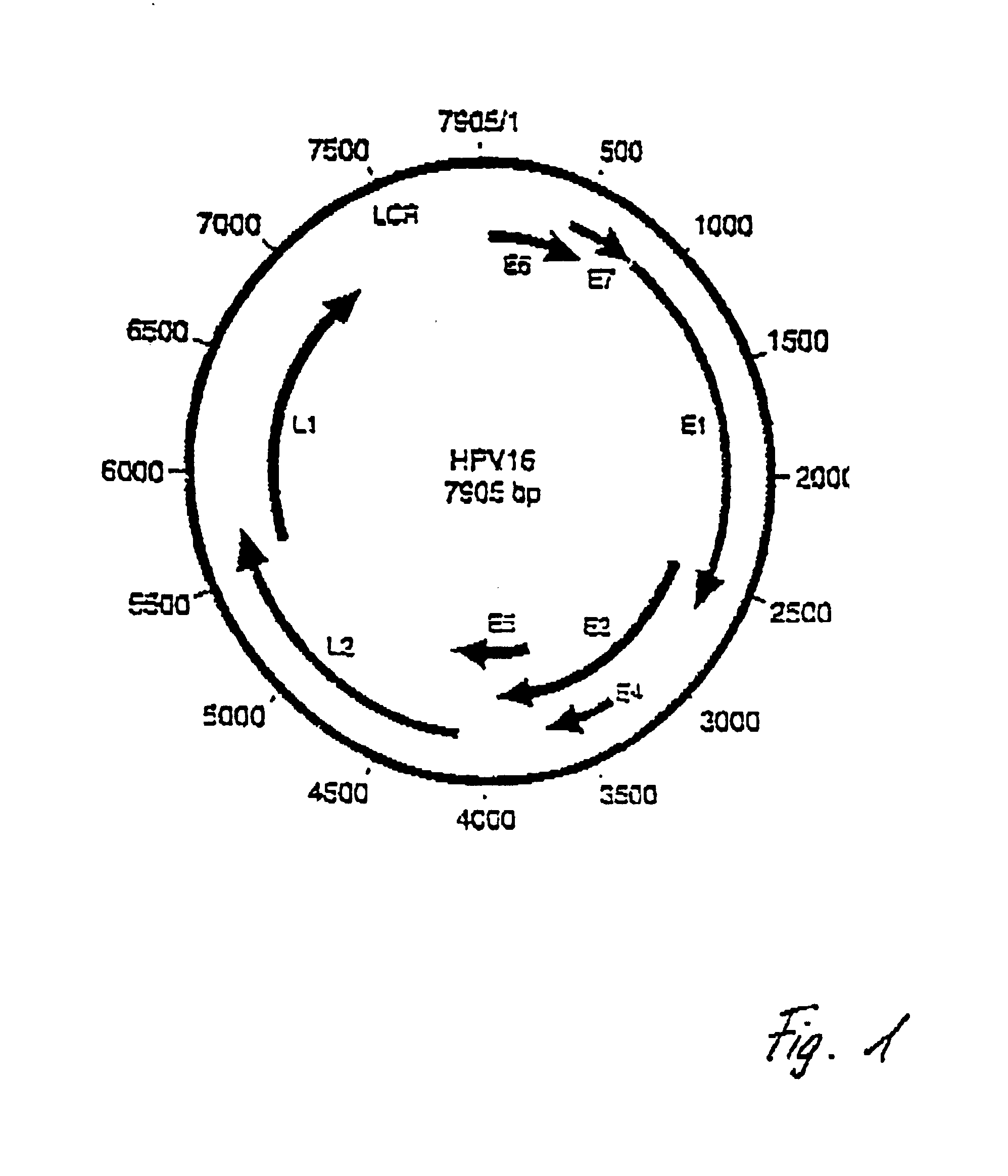
Detection of human papillomaviruses
a human papillomavirus and detection method technology, applied in the field of human papillomavirus detection, can solve the problems of low detection efficiency, low detection efficiency, and inability to reliably detect all hpv viruses
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
DNA Extraction
[0054]For the purpose of detecting HPV, it is first of all necessary to remove biopsy or smear material. This is taken up in 500 μl of melting buffer (0.1 M EDTA; 0.05 M Tris, pH 8.0; 0.5% SDS). 2.5 μl of proteinase K (20 mg / ml) are then added to this and the solution is incubated at 55° C. for from 24 to 48 hours.
[0055]This is then followed by two extractions with phenol (phenol:CHCl3:isoamyl alcohol=25.24:1). The preparation is mixed, not vortexed, incubated for 2-5 min. (rotated with inversion), and, after that, centrifuged at 14 000 rpm for 5 min. There then follows an extraction with CHCl3 / isoamyl alcohol (24:1) and incubation for 2-5 min and, after that, a centrifugation at 14 000 rpm for 5 min.
[0056]{fraction (1 / 10)} of the preparation volume of 7.5 M ammonium acetate is then added and the preparation is mixed and incubated at 55° C. for from 2 to 4 hrs.
[0057]The preparation is then precipitated with ethanol at −20° C. overnight.
[0058]The DNA which has been e
example 2
PCR Reaction
[0059]The PCR is used for selectively amplifying viral DNA segments.
[0060]0.25 μl, 1 μl, 2.5 μl and 5 μl of the extracted quantity of DNA are used for this purpose (without the concentration being determined); additions: primer 1: 100 ng, primer 2; 100 or 200 ng, dNTPs: in each case 10 mM; 5 μl of Taq polymerase buffer (10-fold, without MgCl2); 7 μl of MgCl2 (25 mM stock solution, corresponds to 3.6 mM); 0.4 μl of Taq Gold polymerase (corresponds to 2 units). The preparation is made up to a total volume of 50 μl. The PCR is performed in an MJ-Research Thermocycler (PTC 200); set heating rate: 1° C. / sec. The PCR amplification takes place over 40 cycles [10 min. 95° C. (heating, only in the first cycle); 30 sec. 95° C.; 30 sec. 55° C.; 1 min. 72° C.; 5 min. 72° C. (cooling, only in the last cycle)].
[0061]For the purpose of nonspecifically detecting an HPV infection (screening), preference is given to using the sequences SEQ ID No. 1 (forward) and SEQ ID No. 2 (backward) as
example 3
DNA Sequencing (Typing)
[0067]The sequencing of the amplificate from Example 2, for the purpose of identifying the HPV type, is carried out using the ABI Big Dye Terminator Cycle Sequencing Ready Reaction Kit containing Ampli-Taq and using 200 ng of primer SEQ ID No. 1 or SEQ ID No. 3 and the PCR amplification preparation. This latter has previously been concentrated and purified through Microcon 100 columns (Millipore). Approx. 50% of the purified preparation is used for the sequencing.
[0068]The sequencing PCR reaction is carried out in accordance with the following conditions: 10 sec. 96° C., 5 sec. 55° C., 4 min. 60° C., 30 cycles, with a heating rate of 1° C. / sec.
[0069]There then follows a precipitation with 0.3 M sodium acetate and 2.5 times the volume of ethanol.
[0070]The pellet is taken up in 25 μl of TSR (template suppression reagent from ABI), incubated at 90° C. for 2 min. and, after that, brought to 4° C. for 2 min. and subsequently kept at room temperature.
[0071]The capill
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap