Escherichia coli camp receptor protein mutant, genetic engineering strain and application
A technology of genetically engineered bacteria and Escherichia coli, applied in the field of genetic engineering, can solve the problems such as the need for further improvement of xylitol concentration and production efficiency, insufficient transport and conversion rate of xylose, environmental pollution, etc. The effect of reducing equipment investment and reducing sewage discharge
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Taking IS5-5 as the starting strain, the steps to construct the IS5-5G strain (I112L, T127G, and A144T three-site mutations of the cyclic AMP receptor protein CRP) using the CRISPR / Cas9 method are as follows:
[0053] 1) Construction of pTarget-G plasmid
[0054] Firstly, it is necessary to find a PAM site on the target gene, determine the corresponding N20 sequence, and replace the cadAspacer on the pTargetF plasmid with the N20 sequence of the target gene. Using N20-crp-F / N20-crp-R as primers and pTargetF plasmid as template, the pTarget-G plasmid was constructed by PCR of the whole plasmid mutation.
[0055] 2) Fix template building
[0056] Using the Escherichia coli W3110 genome as a template, CrpG-u-F and CrpG-u-R as primers to amplify the upstream homology arm, and CrpG-d-F and CrpG-d-R as primers to amplify the downstream homology arm, run nucleic acid electrophoresis and recover with related kits. Then, using CrpG-u-F and CrpG-d-R as primers and an equal pro
Embodiment 2
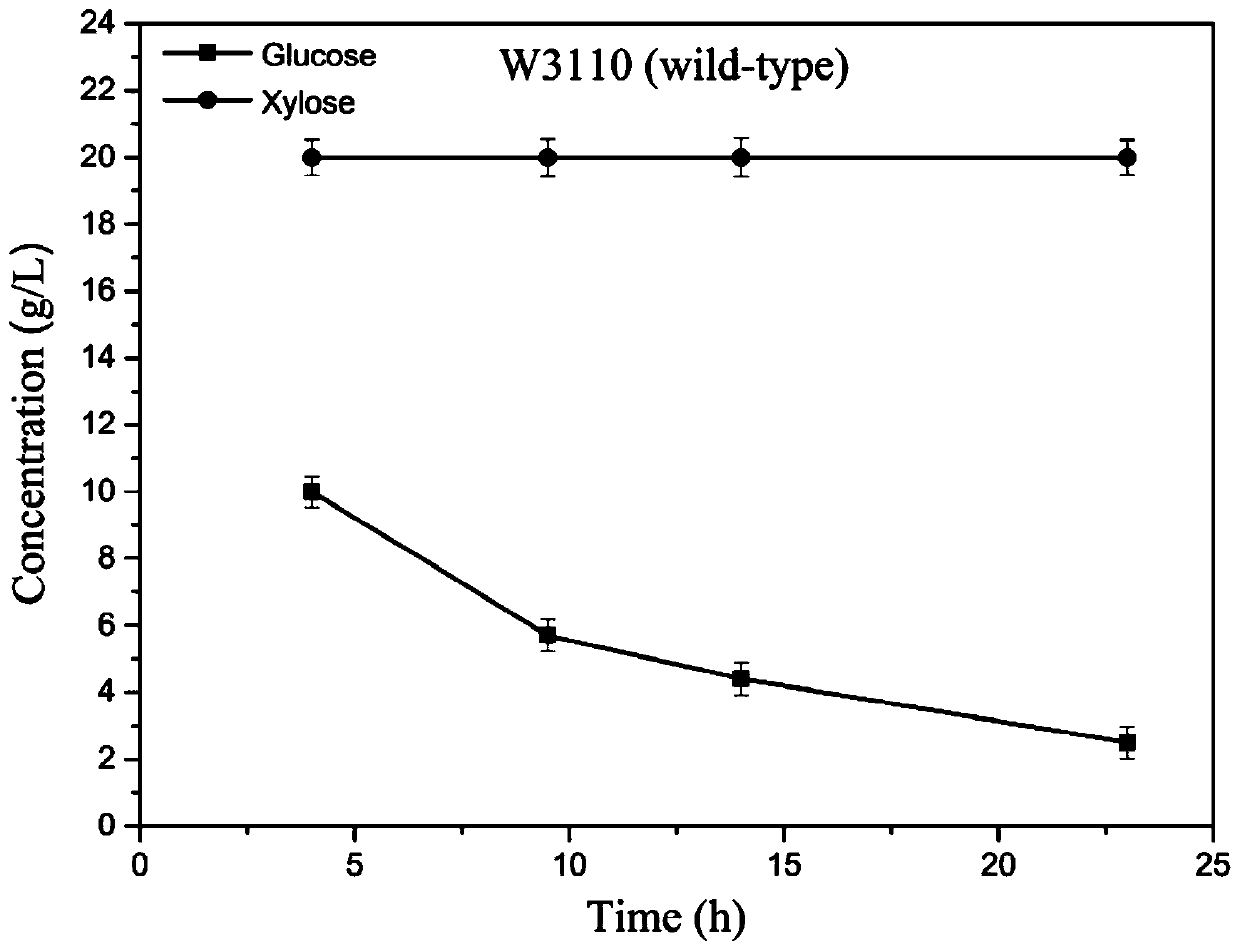
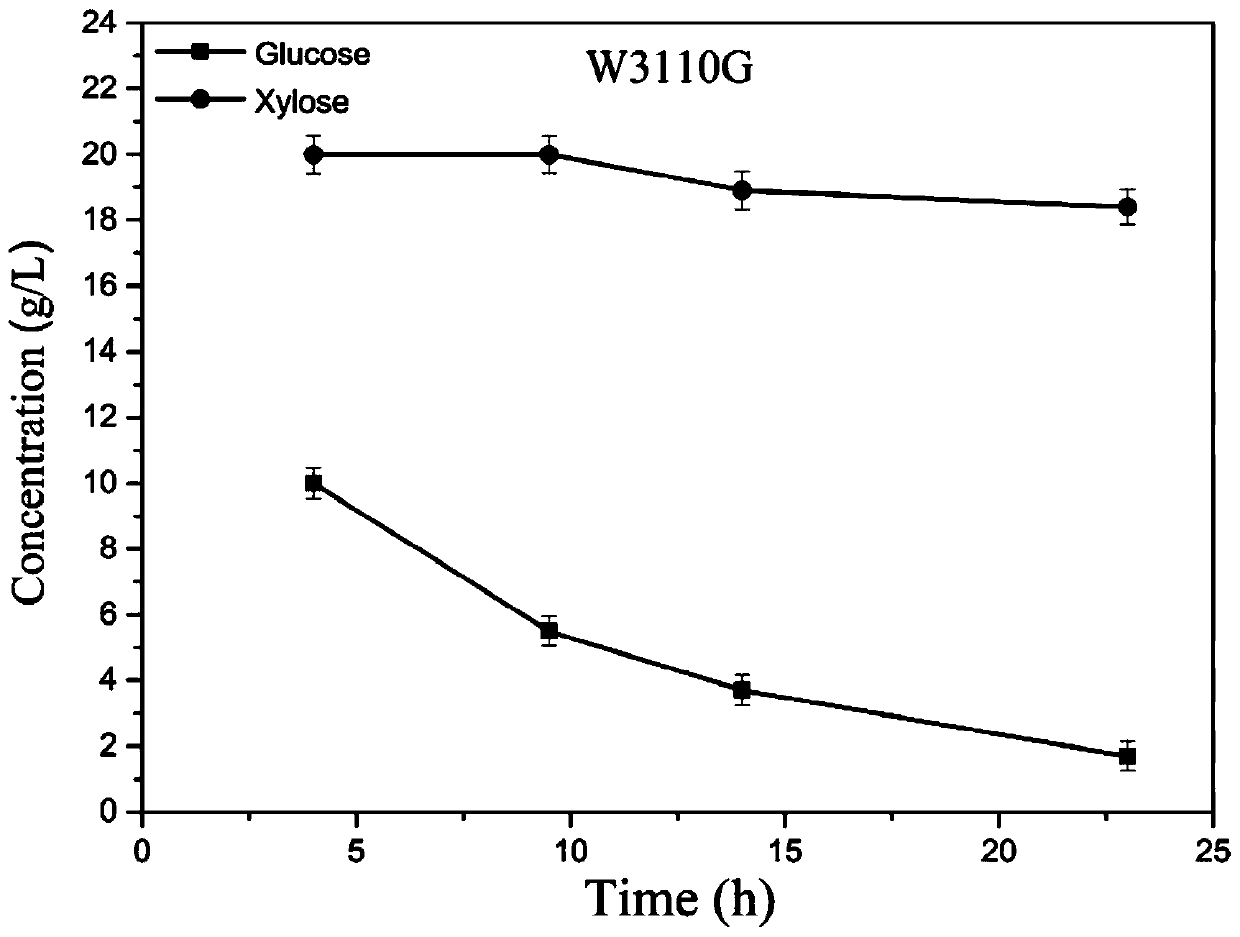
[0079] Using the W3110 strain as the starting strain, the CRISPR / Cas9 method was used to construct the W3110G strain (the I112L, T127G, and A144T three-site mutations of the cyclic AMP receptor protein CRP).
[0080] When using the CRISPR / Cas9 method to construct, the method is the same as in Example 1, and the primers and plasmids used are the same as the pTarget-G plasmid and repair template in steps (1) and (2) in Example 1.
Embodiment 3
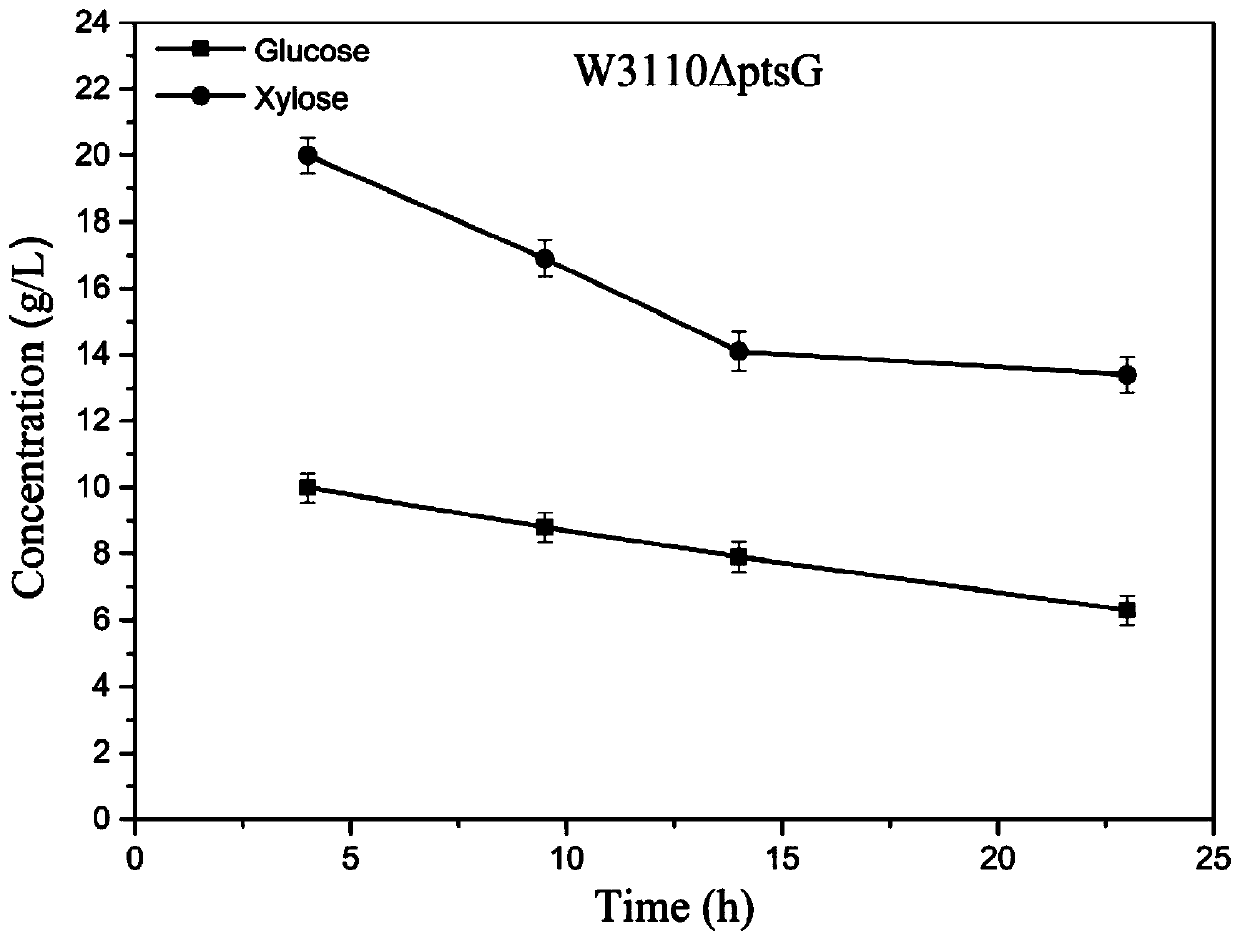
[0082] Using Escherichia coli W3110 as the starting strain, the CRISPR / Cas9 method was used to construct the W3110ΔptsG strain (knockout the ptsG gene), and the steps were as follows:
[0083] Using N20-ptsG-F / N20-ptsG-R as primers and pTargetF plasmid as template, the whole plasmid mutation PCR was used to construct pTarget-ptsG plasmid. Using the Escherichia coli W3110 genome as a template, Del-ptsG-u-F and Del-ptsG-u-R as primers to amplify the upstream homology arm, Del-ptsG-d-F and Del-ptsG-d-R as primers to amplify the downstream homology arm, run nucleic acid Electrophoresis and recovery with relevant kits. Then, using Del-ptsG-u-F and Del-ptsG-d-R as primers and an equal ratio mixture of the upstream and downstream homology arms as templates, overlap extension PCR was performed, and fragments of corresponding length were recovered by gel cutting to obtain a repair template for knocking out the ptsG gene.
[0084] The primers used for strain construction are listed in
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap