Construction method of monascus ruber for producing lovastatin in high yield instead of citrinin
A technology of Monascus ruberus and lovastatin, which is applied in the field of bioengineering and can solve single problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
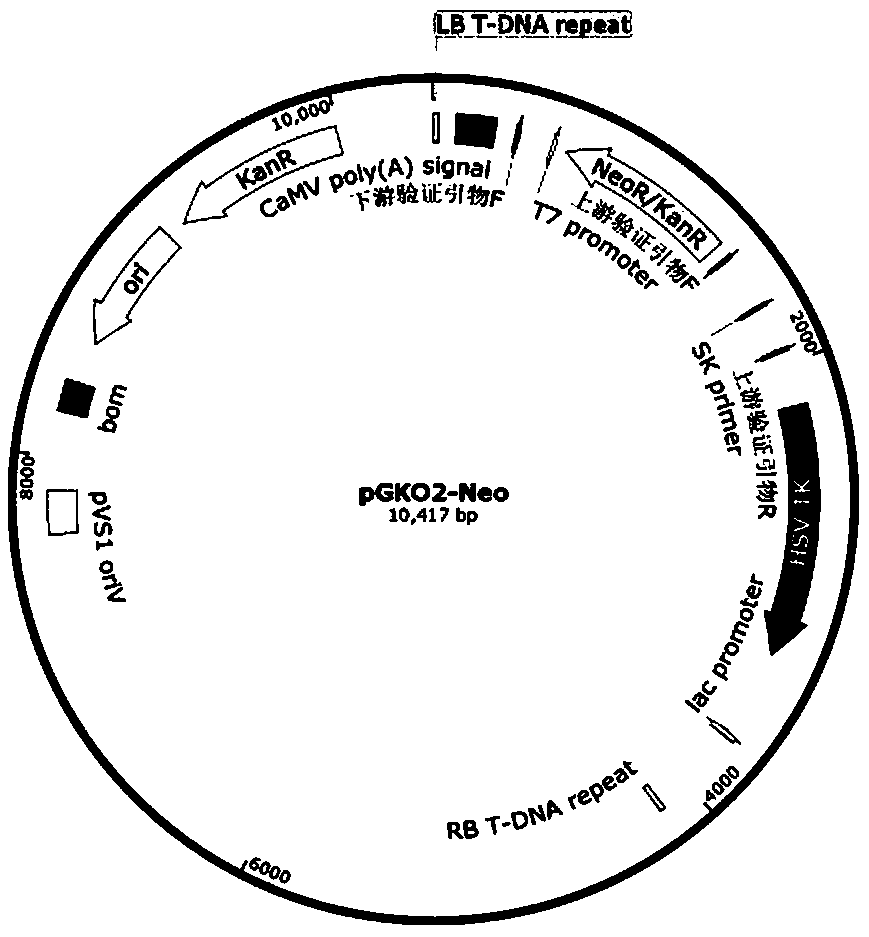
[0045] Example 1 Construction of GN-01△ ctnR Knockout plasmid (pGKO2-Neo-up- ctn R-dn)
[0046] (1) design ctnR One pair of upstream and downstream homology arm primers
[0047] ctnR -up-F::5'-GGGGTACCGGTACCTTGAGGGAATGGAGC-3' (kpnI);
[0048] ctnR -up-R: 5'-ACGCGTCGACGTACTGCAGACATCGGCGAG-3' (SalI);
[0049] ctnR -dn-F:5'-CGGGATCCTAACTAGGAAACGCGCAGGTC-3'(BamHI);
[0050] ctnR -dn-R: 5'-GCTCTAGACAAAGTAATCGACGATGTGCG-3' (XbaI);
[0051] (2) Amplified by PCR ctnR upstream and downstream homology arms
[0052] PCR amplification system: 10×TransStart FastPfu Buffer 5 μL, High Pure dNTPs (2.5 mmol / L) 4 μL, DNA template 1 μL, primer F 1 μL, primer R 1 μL, TransStart FastPfu DNA Polymerase 1 μL, ddH 2 O to make up to 50 μL.
[0053] PCR reaction parameters: pre-denaturation at 98°C for 2 min; denaturation at 98°C for 15 s, annealing at 55°C for 15 s, extension at 72°C for 1 min, 35 cycles; incubation at 72°C for 5 min; storage at 4°C.
[0054] The DNA sequence dete
Embodiment 2
[0060] Transformation of embodiment 2 GN-01 bacterial strain
[0061] The constructed knockout plasmid (pGKO2-Neo-up- ctn R-dn) was electroporated into Agrobacterium (AGL-1), and Agrobacterium transformants containing knockout plasmids (AGL-1- pGKO2-Neo- up- ctn R-dn). Agrobacterium containing the knockout plasmid was cultured in induction medium (IM) containing 200 μmol / L acetosyringone for 9-10 h, OD 600 is about 0.5, and Monascus ruber GN-01 spores (concentration 10 7 / mL) were mixed in equal volume, spread on the symbiotic medium containing acetosyringone, and cultured at 25°C in the dark for 2-3 days. The hyphae on the symbiotic plate were washed, spread on a PDA plate containing 300 μg / mL cefotaxime and 50 μg / mL neomycin, and cultured at 30°C for 4-5 days to obtain knockout transformants. Then pick the transformants on the PDA re-screening plate containing 300 μg / mL cefotaxime and 50 μg / mL neomycin to form a single colony and inoculate liquid PD medium,
Embodiment 3
[0062] Example 3 GN-01△ ctnR Knockout strain validation
[0063] (1) Design primers for PCR verification
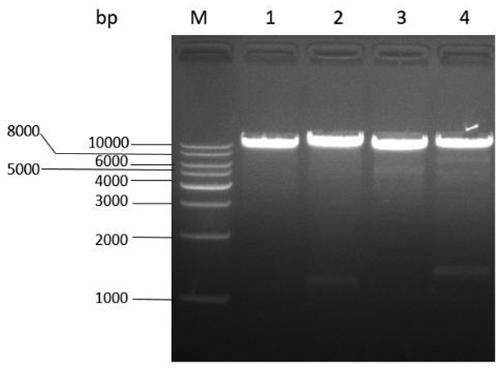
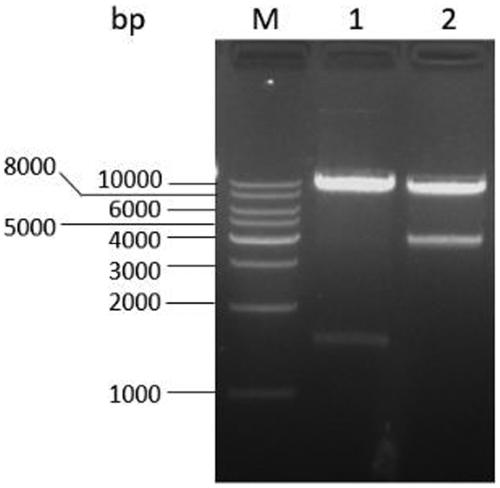
[0064] The transformants grown on the neomycin-resistant plate were continuously passed on for 5 generations, and the transformants with stable inheritance were screened out. Extract the genomic DNA of the transformant and use it as a template to design primers on the upper and lower homology arms ctnR -F and ctnR -R; in ctnR Design within the gene deletion region△ ctnR -F and △ ctnR -R primers for PCR verification. If primer ctnR -F and ctnR -R, the band amplified by the transformed strain is about 1500 bp, while the band amplified by the control is about 1200 bp ( Figure 5 ); with primer △ ctnR -F and △ ctnR -R, the control strain can amplify a band of about 900 bp, but the transformed strain cannot amplify the band ( Image 6 ), it is preliminarily proved that the transformant has successfully carried out the replacement-type homologous recombinatio
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap