Tools for the identification of lingo-1, lingo-2, lingo-3 and lingo-4 ligands, and uses thereof
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
Lingo-1 Forms a Dimer In Vivo
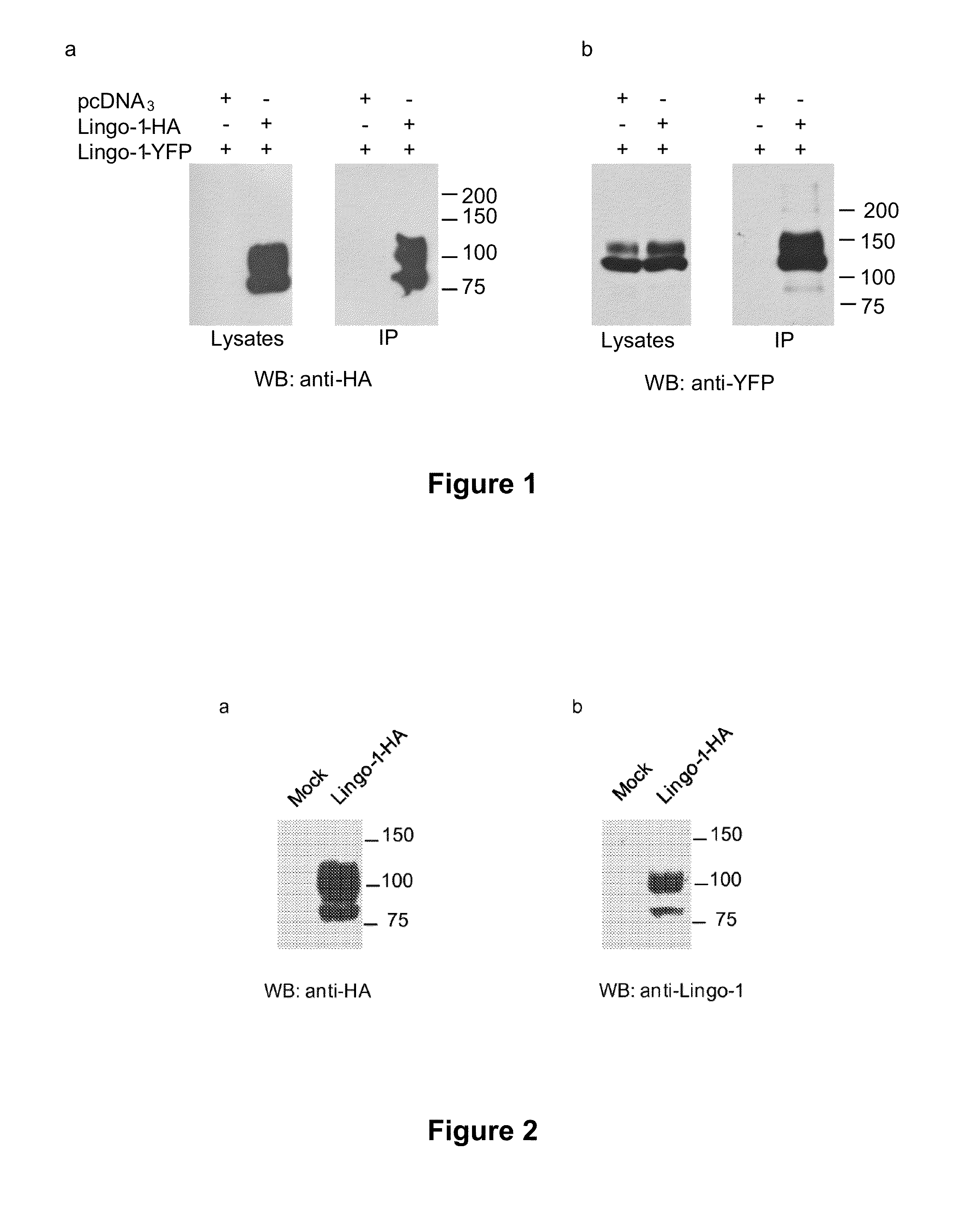
1 / Lingo-1 Forms a Dimer in HEK-293 Cells
[0130]HEK-293 cells were first of all cotransfected with two plasmids which allow the expression of the Lingo-1 protein tagged with HA and Lingo-1 tagged with a fluorescent protein, YFP. Communoprecipitation experiments showed that, when the HA-tagged Lingo-1 protein is immunoprecipitated with agarose beads coupled to an anti-HA antibody (FIG. 1a), the YFP-tagged Lingo-1 protein is coimmunoprecipitated (FIG. 1b). These results show, for the first time, that Lingo-1 formed a dimer in vivo. These results also show for the first time that the whole and membrane form of Lingo-1 was capable of forming dimers in HEK-293 cells.
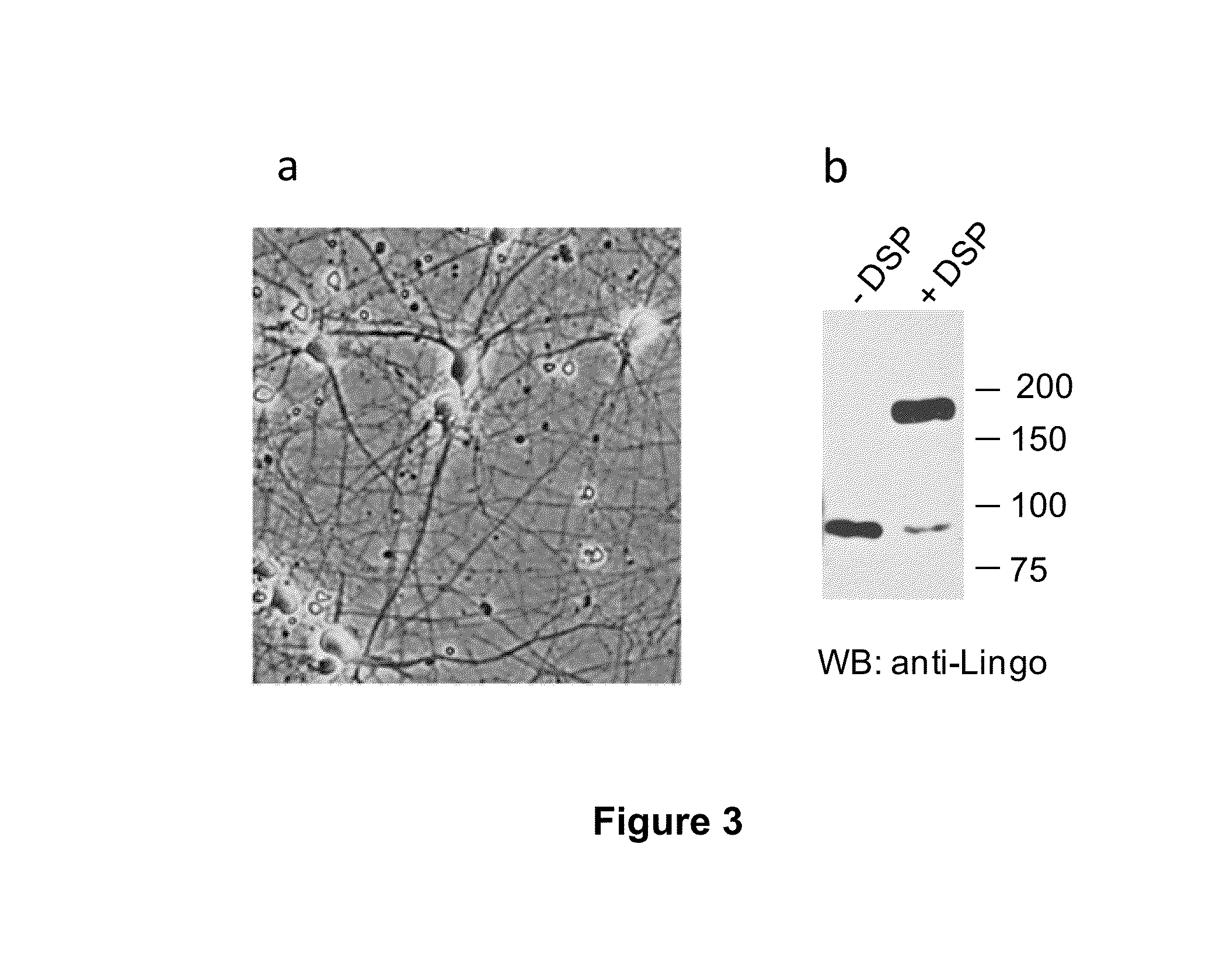
2 / Lingo-1 Forms a Dimer in Neurons
[0131]After validation of the anti-Lingo-1 antibody (FIG. 2), existence of dimers in cortical neurons in culture was tested (FIG. 3a). Given that certain detergents used during the lysis can induce the formation of protein aggregates and result in the artefactual f
example 2
Detection of the Oligomerization of the Lingo-1 Protein in Live Cells Using the Bret Technique
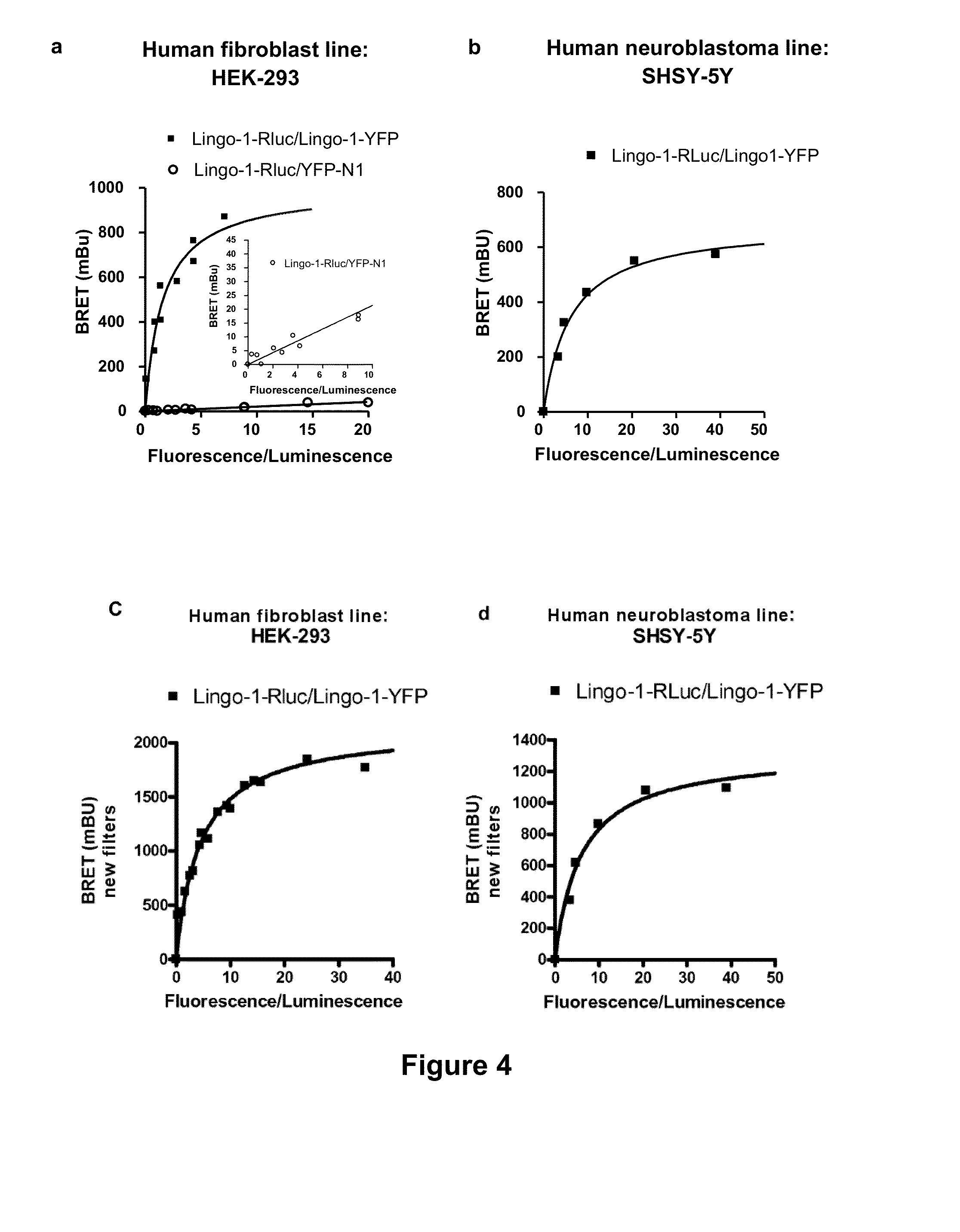
[0133]HEK-293 or SHSY-5Y cells were transfected with a fixed concentration of donor (100 ng of Lingo-1-RLuc plasmid) and increasing concentrations of acceptors, Lingo-1-YFP or the YFP fluorescent protein alone (from 50 ng to 4000 ng of plasmids). The BRET measurement was carried out 48 hours after transfection on live cells using sets of old filters and of new filters.
[0134]The BRET signal observed with the old filters corresponding to the oligomerization of Lingo-1 is clearly specific since, when Lingo-1-RLuc and Lingo-1-YFP were coexpressed, the signal increased according to a hyperbolic curve and reached an asymptote in the two cell types (cf. FIGS. 4a and 4b). Furthermore, when Lingo-1-Rluc was coexpressed with the YFP protein alone, the BRET signal was much weaker (a, insert); thereby attesting to the appearance of nonspecific and random interactions.
[0135]These results show that the Ling
example 3
The Interaction Between Lingo-1-RLuc and Lingo-1-YFP is Specific and can be Modulated
[0137]HEK-293 cells were transfected with the Lingo-1-RLuc and Lingo-1-YFP fusion proteins (the donor / acceptor ratio remains constant) and increasing concentrations of the whole Lingo-1 protein (Lingo-1-HA, amino acids 1-620) or the dominant negative thereof (DN-HA, amino acids 1-577) tagged with HA. The BRET measurement was carried out 48 hours after the transfection on live cells. The HA-tagged constructs significantly decreased the BRET signal obtained between Lingo-1-RLuc and Lingo-1-YFP (p<0.001 vs control, ANOVA, NewMan Keuls). The results are shown in FIG. 5.
[0138]The BRET signal obtained between the Lingo-1-RLuc and Lingo-1-YFP proteins is therefore specific and can be modulated. Indeed, when the HA-tagged whole Lingo-1 protein (620 amino acids) or the HA-tagged dominant negative of Lingo-1 (577 amino acids, without the cytoplasmic portion of the protein) was overexpressed at 800 ng, the BRET
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap