Assay for Screening of Anti-Viral Compounds That Inhibit Specific Interaction Interfaces Between Cullin5 and an ElonginB/ElonginC/ CBF-beta/HIV-1 Vif Complex
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Example
Example 1
Core-Binding Factor β (CBFβ) Increases the Affinity Between Human Cullin 5 and HIV-1 Vif within an E3 Ligase Complex
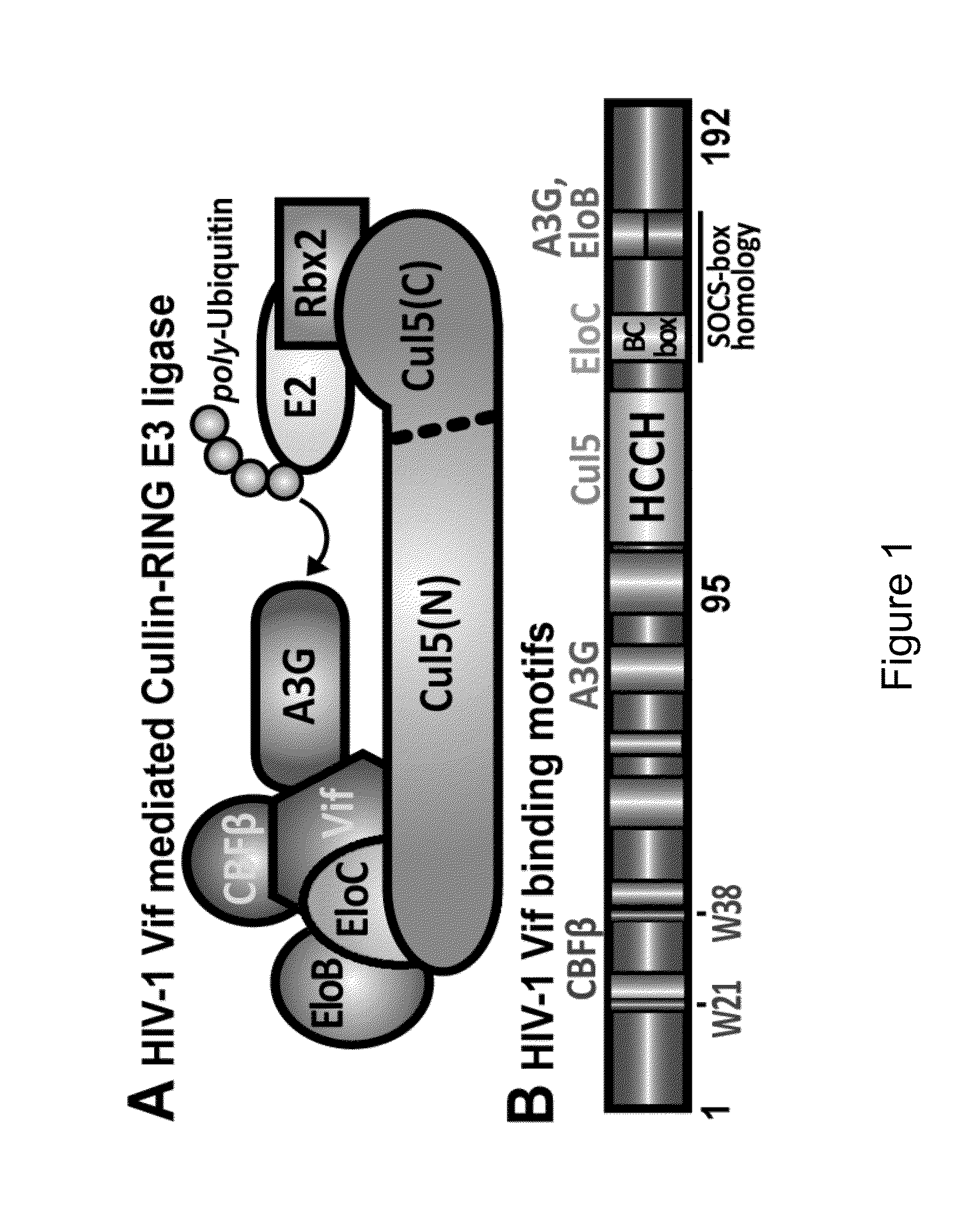
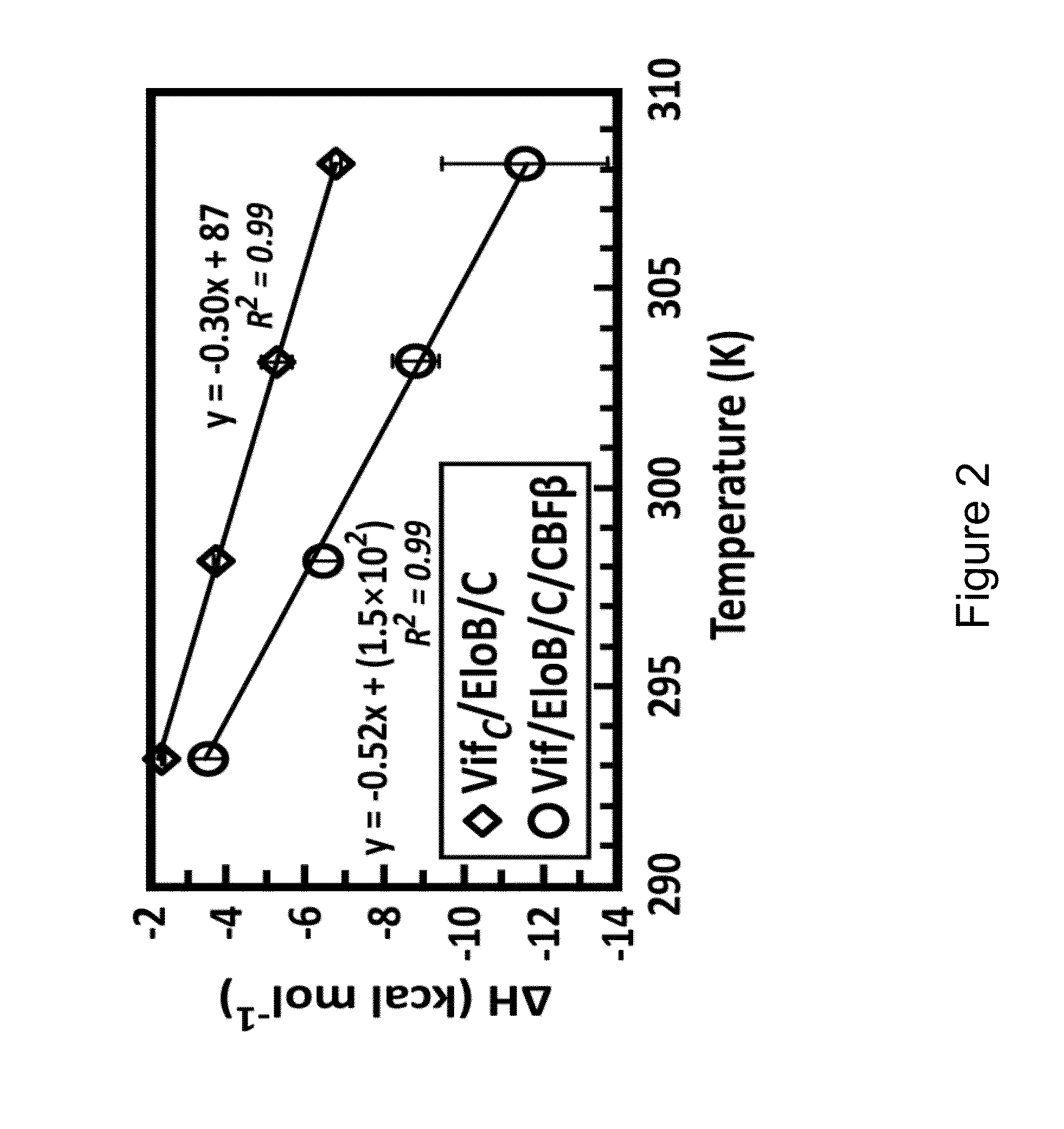
[0244]HIV-1 Vif masquerades as a receptor for a cellular E3 ligase harboring ElonginB, ElonginC, and Cullin5 (EloB / C / Cul5) proteins that facilitate degradation of the antiretroviral factor A3G. This Vif-mediated activity requires human CBFβ in contrast to cellular substrate receptors. As presented herein, it was calorimetrically observed that Cul5 binds tighter to full-length Vif(1-192) / EloB / C / CBFβ (Kd=5±2 nM) than Vif(95-192) / EloB / C (Kd=327±40 nM), which cannot bind CBFβ. A comparison of heat-capacity changes supports a model wherein CBFβ prestabilizes Vif(1-192) relative to Vif(95-192), consistent with a stronger Cul5 interaction with Vif's C-terminal Zn2+-binding motif. The data presented herein suggests that an additional interface between Cul5 and an N-terminal region of Vif may exist, which has therapeutic-design implications.
[0245]The materia
Example
Example 2
Novel Sequences in HIV-1 Vif and Human Cullin5 that Mediate Vif-Cullin5 Binding
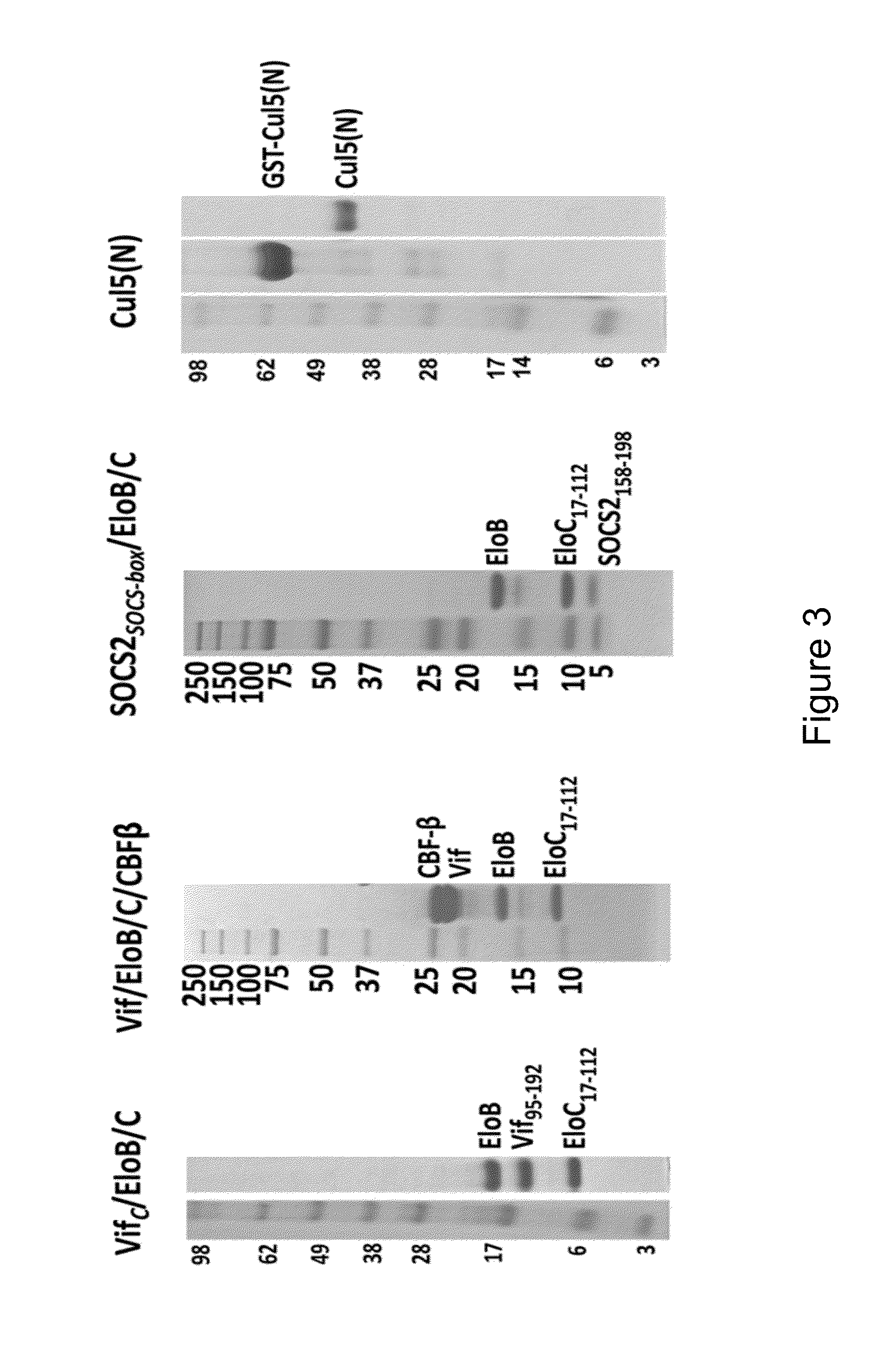
[0270]Described herein is the discovery of novel sequences in HIV-1 Vif and human Cullin5 (Cul5) that have not been previously described but are now shown to be implicated in forming portions of the molecular interaction interface between Cul5 and the essential HIV-1 protein Vif. As discussed elsewhere herein, Vif is embedded as an integral, full-length sequence component of a multi-protein complex comprising ElonginB / ElonginC / Vif / CBFβ, which is part of a larger human host Cullin-RING E3 ubiquitin-ligase complex whose biological role is to degrade innate immune factors of the host such as APOBEC3G. Knowledge of unique interacting peptides between the host and virus provides a significant advantage in efforts to develop peptide-like molecules intended to disrupt protein interfaces that are essential for viral infectivity.
[0271]The HIV-1 Vif residues that undergo dynamic exchange upon binding Cu
Example
Example 3
Binding of Peptide Inhibitors to Vif / EloB / C / CBFβ
[0276]As described elsewhere herein, peptide sequences (SEQ ID NOs: 9-11) were identified that appear to be present in the interface between human Cul5(N) and the HIV-1 Vif / EloB / C / CBFβ complex. Using these results, specific peptides were designed and studied for the ability to bind to the isolated HIV-1 Vif / EloB / C / CBFβ complex. The data presented herein demonstrates the construction of a “lead platform” for the generation of peptides or derivatives thereof that will block the HIV-1 Vif interaction with the host, thereby eliminating an essential viral interaction.
[0277]As described elsewhere herein, the following sequences of Cul5 were identified by HDX-MS to participate in the binding between Vif and Cul5: (A) 30LRQESVTKQQW40 (SEQ ID NO: 9), (B) 43LFSDVHAVCL52 (SEQ ID NO: 10), and (C) 55DKGPAKIHQAL65 (SEQ ID NO: 11). Using this information, peptides that comprise one or more regions of A, B, or C, or fragments t
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
| Antimicrobial properties | aaaaa | aaaaa |
| Fluorescence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap