Engineering bacterium strain and method for preparing farnesene by using cellulose
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] This example is used to illustrate the preparation of cellulose hydrolyzate.
[0039] Weigh 100 g of corn stalks with a particle size of 20-80 meshes, add 1.5 L of formic acid solution (containing 88 wt % formic acid and 1 wt % HCl) and place it in a treatment tank. After reacting in a constant temperature water bath at 65° C. for 3 h, the reaction slurry was separated from solid to liquid. The solid component is further treated with ammonia water, the treatment conditions are: the concentration of ammonia water is 15wt%, the volume of ammonia water per kilogram of solid component is 8 liters, the treatment temperature is 60° C. and the reaction time is 16 hours. After the reaction, the material is separated from the solid and liquid, the solid component is washed with distilled water to pH 6-7, and dried to obtain the pretreated solid material.
[0040] The pretreated solid material is subjected to enzymatic hydrolysis, and the enzymatic hydrolysis conditions include: th
Embodiment 2
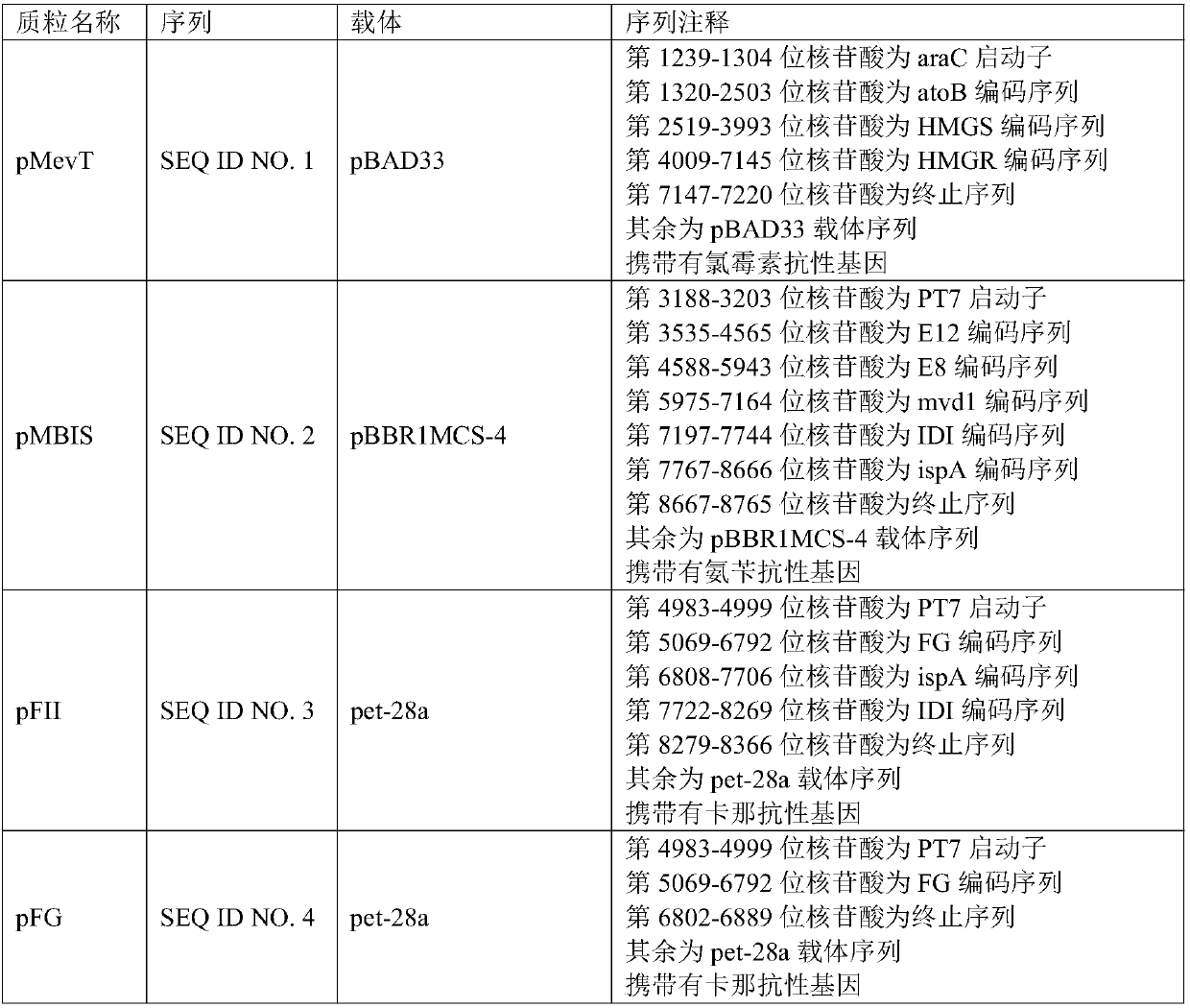
[0042] This example is used to illustrate the sequence structure of the plasmid. Commercially available vectors used in this example include pBAD33 vector, pACYDCDuet-1 vector, pBBR1MCS-4 vector and pet-28a vector.
[0043] According to the instruction manual of the above-mentioned vectors, a set of plasmids for the metabolic pathway of MVA was constructed by using molecular biology operations such as PCR cloning, whole gene synthesis, restriction endonuclease digestion and ligation, including pMevT plasmid, pMBIS plasmid and pFII plasmid, and then One plasmid, the pFG plasmid, was constructed to replace the pFII plasmid, followed by the construction of a pMevT-1 plasmid to replace the pMevT plasmid. The sequence information of the above plasmids is specifically shown in Table 2.
[0044] Table 2
[0045]
Embodiment 3
[0047] This example is used to illustrate the construction process of engineering strains.
[0048] Construction of the engineering strain F1 of the experimental group: Escherichia coli BL21 was selected as the host cell, and CaCl 2 The transformation method was transferred to the pMevT plasmid, pMBIS plasmid and pFII plasmid described in Table 2. The strain can synthesize farnesene through the MVA metabolic pathway, and has an optimized and efficient balanced metabolic pathway. On the antibiotic resistance screening plate, select 9 clones of the strain, namely F1-1, F1-2, F1-3, F1-4, F1-5, F1-6, F1-7, F1-8 and F1-9.
[0049] The construction of the engineering strain DF1 of the control group: Escherichia coli BL21 was selected as the host cell, and CaCl 2 The transformation method was transferred to the pMevT plasmid, pMBIS plasmid and pFG plasmid described in Table 2. The strain can synthesize farnesene through the MVA metabolic pathway, but the balanced metabolic pathway h
PUM
| Property | Measurement | Unit |
|---|---|---|
| Particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap