Fluorescent proteins
a fluorescent protein and protein technology, applied in the field of new fluorescent proteins, can solve the problems of prolonged excitation period damaging cells and difficult detection, and achieve the effect of improving the usefulness of fluorescent proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of GFP Plasmids
[0075]Plasmids pEGFP-N1 (GenBank accession number U55762) and pEGFP-C1 (GenBank accession number U55763) both contain a derivative of GFP in which one extra amino acid has been added at position two to provide a better translational start sequence (a Kozak sequence) and so the total number of amino acids is increased by one to 239 instead of the 238 found in wildtype GFP. Therefore the denomination of mutations in GFP in these plasmids strictly should be referred to as e.g. F65L rather than F64L. However, to avoid this source of confusion and because the GFP community has adopted the numbering system of wildtype GFP in its communications, the numbers used here conform to the commonly used naming of mutations in wildtype GFP. The relevant mutations in this respect are F64L, S65T, and E222G.
[0076]Plasmids pEGFP-N1 and pEGFP-C1 contain the following mutations in the chromophore: F64L and S65T. The codon usage of the GFP DNA sequence has been optimized for expre
example 2
Determination of Spectral Properties of Proteins EGFP and eF64L,E222G
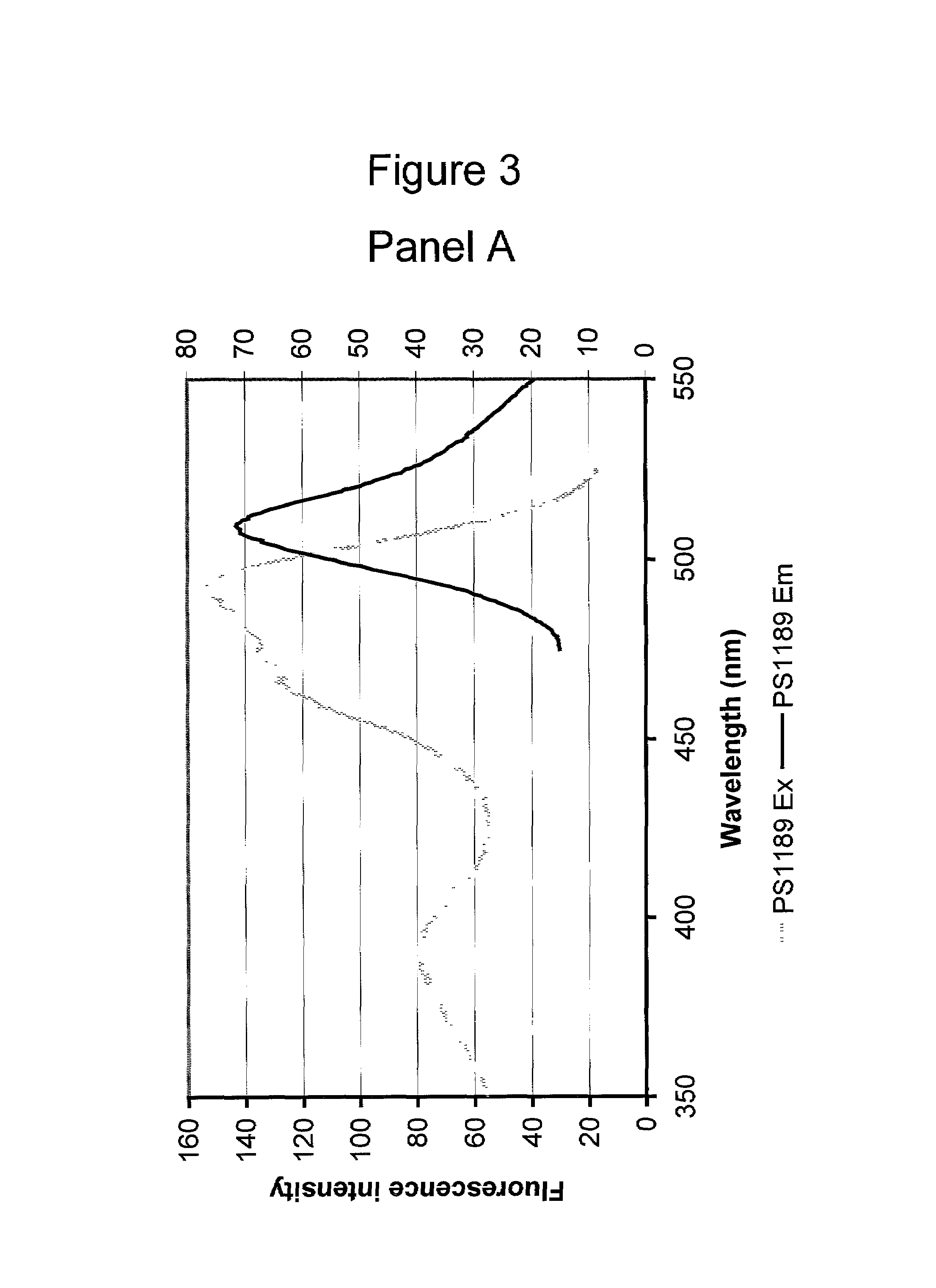
[0092]Plasmids expressing EGFP from plasmid pEGFP-N1 (also referred to as PS279), and eF64L,E222G from plasmid PS699 were transfected into E.Coli TOP10 cells (Invitrogen) using lipofectamine 2000 (from Life Technologies) according to manufacturers recommendations. After 5 days cells were collected and resuspended in extraction buffer 50 mM TRIS(pH 8.0) with 1 mM DTT. Cells were lysed by 3 cycles of freeze-thaw. Cell debris was centrifuged out at 10000 g in acooled centrifuge. NaCl was added to 100 mM.
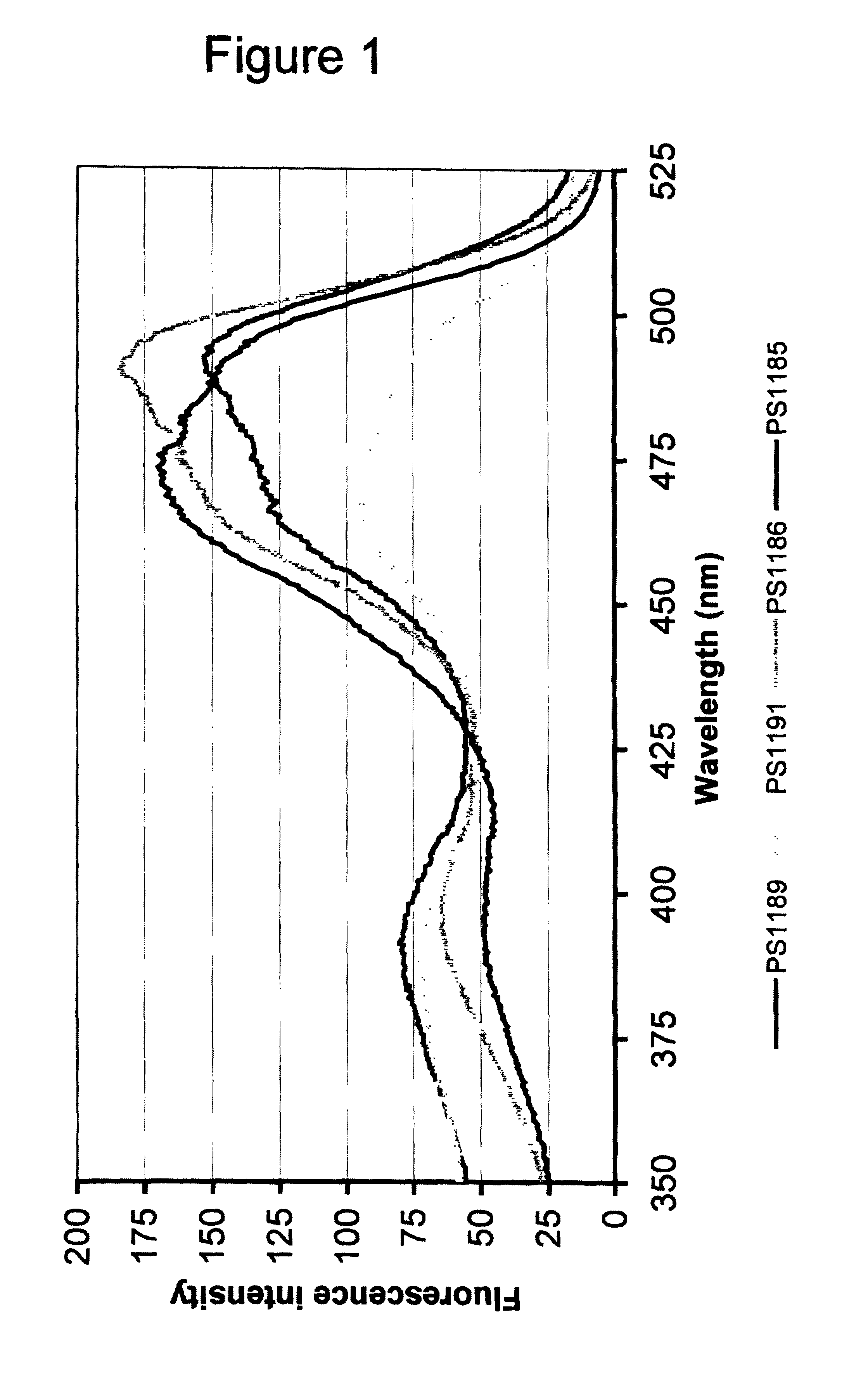
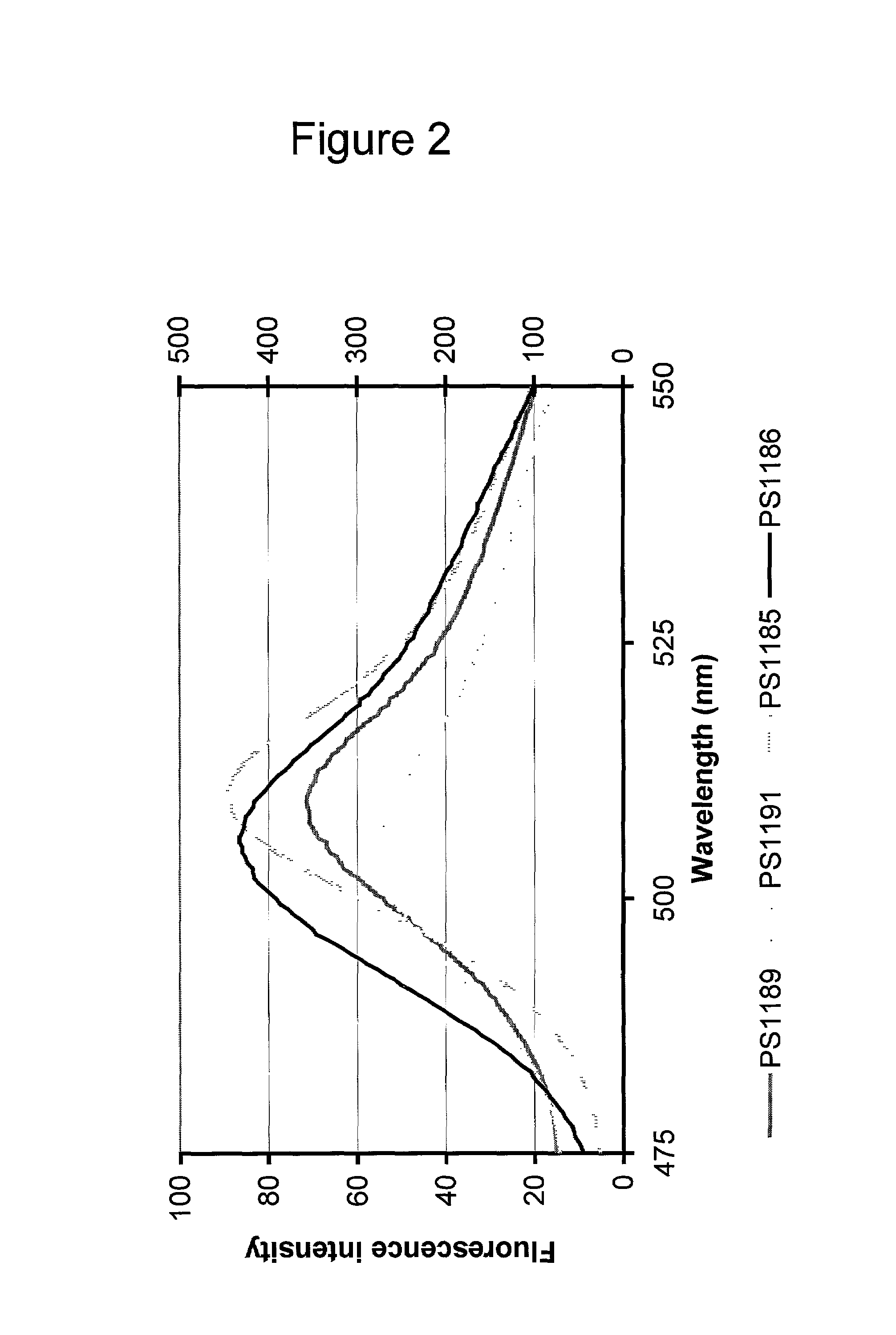
[0093]The cell pellets were resuspended in 1000 μl of H2O each (2-fold dilution relative to volumes of pelleted cultures) and transferred to 1.0×0.5 cm plastic cuvettes and the following excitation and emission spectra were recorded on a Perkin Elmer LS50B luminescence spectrometer:
[0094]
Excitation spectrum:Excitation at 350–525 nm (5 nm slit width) Emission 560 nm (10 nm slitwidth)Data presented in FIG. 1.Emission spectr
example 3
Determination of Time to Fluorescence of EGFP and eF64L,E222G in CHO Cells
[0097]Three, 2 well chambers with CHOhIR cells were transfected with plasmid PS279 expressing EGFP and plasmid PS699 expression eF64L,E222G using the Lipofectamine transfection method.
[0098]Fluorescence from the cells was checked at regular intervals after transfection.
[0099]Lipofectamine 2000 transfection method was used to transfect EGFP and eF64L,E222G in one, 8-well chamber with CHOhIR cells. Fluorescence from the cells was checked at regular intervals after transfection as described above. Images were taken from the same cell fields at each interval. Three different fields were observed for each plasmid. The microscope and camera settings were the same for each image. Optimal exposure time was taken from a chamber of cells with full EGFP expression (transfected 24 hours previously) to ensure the exposure does not saturate. The first images were taken from 45 minutes to 1 hour post transfection, thereafter wi
PUM
| Property | Measurement | Unit |
|---|---|---|
| Time | aaaaa | aaaaa |
| Width | aaaaa | aaaaa |
| Width | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap