Tag removal from proteins expressed in pro- and eukaryotic hosts
a technology of proand and eukaryotic hosts, applied in the field of tag removal from proteins expressed in proand eukaryotic hosts, can solve the problems of ubl-specific proteases in eukaryotic systems that are typically hampered in application, and neither scatg8 fusion proteins nor proteases are well behaved in terms of solubility and/or expression levels, so as to promote the expression and solubility of proteins fused, and achieve high-
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Expression of xlLC3B-Fusions in E. coli
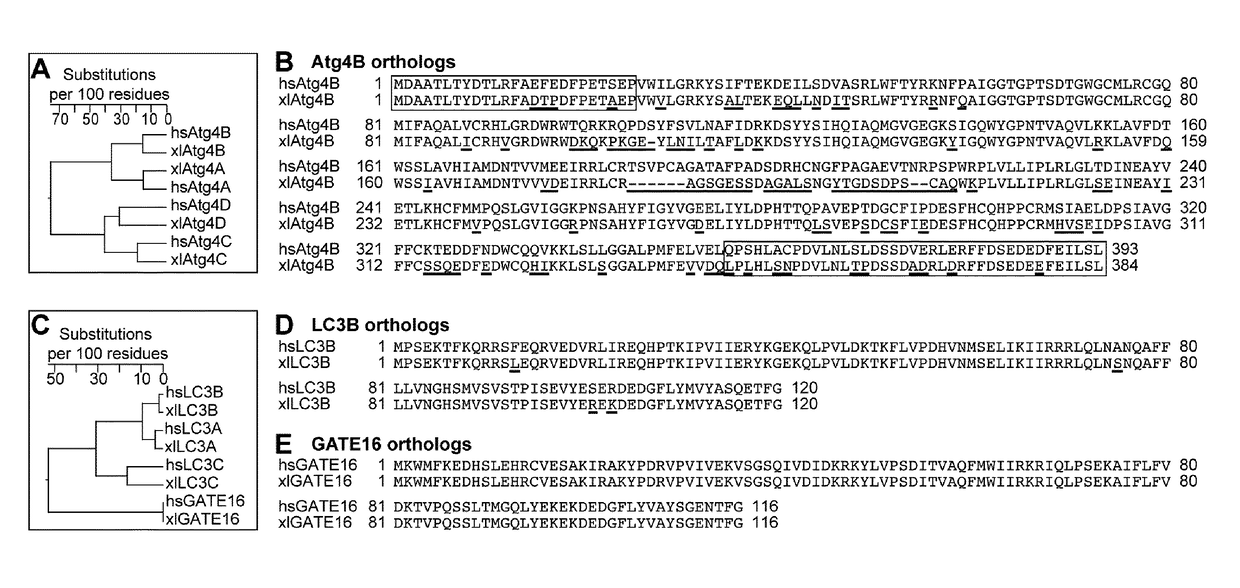
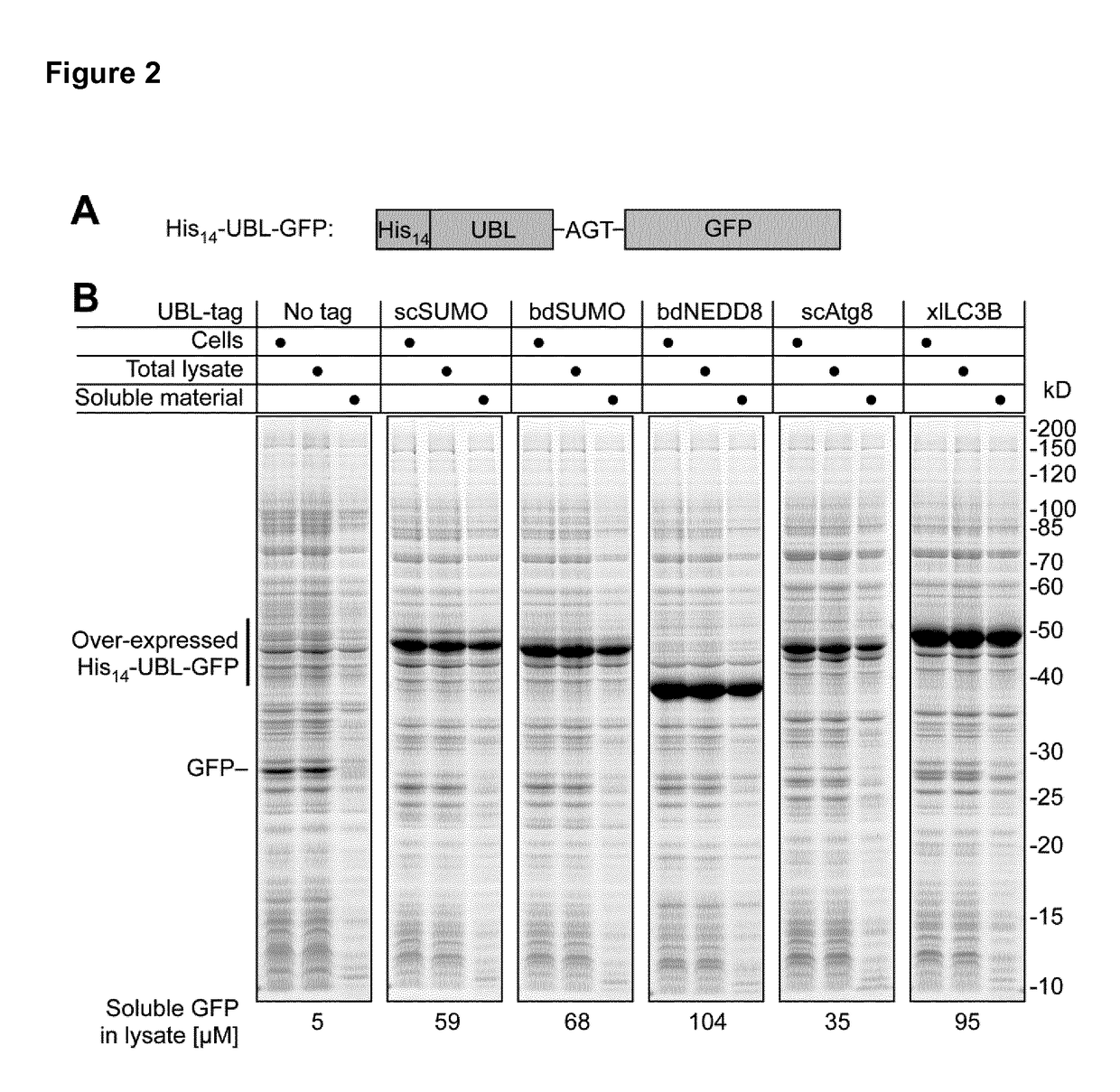
[0222]Initially, the primary aim was to analyze the suitability of xlAtg4B for tag removal from recombinant proteins fused to Xenopus laevis Atg8 orthologs. As the inventors had observed before that fusions to S. cerevisiae Atg8 only show suboptimal expression levels (Frey, S. and Görlich, D. (2014) J Chromatogr A 1337, 95-105), the inventors first compared the impact of various UBLs including xlLC3B on expression and solubility of GFP (FIG. 2). Indeed, xlLC3B-GFP could be highly over-expressed in E. coli and produced nearly 3-times higher levels of soluble GFP as compared to the corresponding scAtg8 fusion. Remarkably, with regard to the expression level, both xlLC3B and bdNEDD8 clearly outperformed scSUMO, which is well known for its expression- and solubility-enhancing effects.
example 2
Identification and Characterization of xlAtg4B Protease and xlAtg4B Protease Fragments
[0223]As a next step, the inventors wanted to find well-expressible and well-soluble xlAtg4B fragments displaying optimal stability and catalytic properties. Based on known structures of the human Atg4B homolog (Kumanomidou, T., Mizushima, T., Komatsu, M., Suzuki, A., Tanida, I., Sou, Y. S., Ueno, T., Kominami, E., Tanaka, K. and Yamane, T. (2006) J Mol Biol 355, 612-618; Sugawara, K., Suzuki, N. N., Fujioka, Y., Mizushima, N., Ohsumi, Y. and Inagaki, F. (2005) J Biol Chem 280, 40058-40065; Satoo, K., Noda, N. N., Kumeta, H., Fujioka, Y., Mizushima, N., Ohsumi, Y. and Inagaki, F. (2009) EMBO J 28, 1341-1350), full-length xlAtg4B (residues 1-384) and five shorter xlAtg4B fragments harboring N- and / or C-terminal truncations (xlAtg4B14-384, xlAtg4B25-384 xlAtg4B1-345, xlAtg4B14-345 and xlAtg4B25-345) were cloned and expressed. All proteases fragments could be over-expressed in E. coli and obtained in h
example 3
Application of the xlAtg4B Protease System for Tag Removal and On-Column Cleavage
[0246]An important application of tag-cleaving proteases is on-column cleavage of recombinant proteins. The inventors directly addressed the suitability of xlAtg4B14-384 for this purpose using polyHis-tagged substrate proteins bound to a Silica-based Ni2+ chelate resin of high porosity (FIG. 9). More specifically, ≈100 μM of His14-IF2d1-xlLC3B-GFP or His14-IF2d1-xlGATE16-GFP were immobilized on the respective matrices along with the control protein His14-bdNEDD8-mCherry (FIG. 9A) before incubation with defined concentrations of xlAtg4B14-384 or bdNEDP1 for 1 h at 4° C. Under these conditions, 250-500 nM of xlAtg4B14-384 was sufficient for near-quantitative elution of GFP from the Silica-based resin (FIG. 9B, C). The cleavage was specific as even at much higher concentrations of xlAtg4B14-384 no elution of the bdNEDD8-tagged mCherry control protein could be detected. Vice versa, after treatment with a high
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap